Gene
KWMTBOMO05092 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012515
Annotation
S-adenosylmethionine_synthetase_[Bombyx_mori]
Full name
S-adenosylmethionine synthase
Alternative Name
Methionine adenosyltransferase
MAT
MAT
Location in the cell
Cytoplasmic Reliability : 4.057
Sequence
CDS
ATGCCGGAGACTTCAAAAATGAATGGATACGCGAAAACCAACGGACACAGTTATGATATGGAAGATGGATCAGTATTTTTGTTCACATCGGAATCTGTTGGCGAGGGTCATCCAGACAAAATGTGCGACCAAATAAGCGACGCTATTCTAGACGCGCACCTGAATCAGGATCCGGACGCAAAAGTTGCATGTGAAACCATAACTAAAACCGGTATGGTGCTTTTGTGTGGCGAAATCACATCCAAAGCTAACGTGGATTATCAAAAAGTTGTGCGCGAAACGGTCAAACATATTGGTTATGATGATTCGTCCAAAGGCTTTGATTACAAGACATGCAGTGTGATGCTCGCACTAGACCAACAATCACCAAACATTGCTGCTGGGGTGCATGAGAACAGAAATGACGAGGAAGTTGGGGCAGGAGACCAGGGCTTGATGTTCGGTTATGCAACAGATGAGACAGAAGAATGCATGCCGTTGACTGTAGTGCTTGCACACAAACTCAATCAGAAAATTGCAGAGCTCAGGCGAAATGGAGAATTTTGGTGGGCAAGACCAGATTCAAAAACACAGGTTACTTGCGAATATGTATTTGCTGGTGGTGCAACAGTCCCACAGAGAGTTCATACTGTAGTCGTGTCACTGCAACACTCTGAAAAGATTACCCTAGAGACCTTGCGAGATGAGATCAGAGAAAAGGTCATCAAAGAGGTTATCCCTGCACAATATCTTGATGAGAGAACTGTAATTCACATTAATCCATGTGGACTCTTTATAATTGGAGGACCTCAGTCAGACGCGGGCCTGACGGGACGCAAAATCATCGTGGACACGTATGGCGGGTGGGGGGCGCACGGCGGGGGCGCTTTCTCTGGCAAGGACTTCACCAAGGTCGACCGCTCCGCAGCATACGCCGCCCGATGGGTCGCCAAGTCGCTCGTCAAGGCGGGCTTGTGCCGGCGCTGCATGGTGCAGGTTGCTTATGCTATTGGTGTAGCTGAGCCATTATCGATTACAGTCTTTGATTATGGCACTTCACATAAGACTCAGCAACAACTGCTCTCCATCGTACAAAAGAATTTCGATTTACGACCGGGCAAAATCGTCAAAGAGTTGAACCTCAGAGCACCAATTTATCAGAGAACAAGTACCTATGGACACTTTGGAAGGGCAGGATTCCCCTGGGAGAGTCCCAAGCCGCTGATTGTAGAATGA
Protein
MPETSKMNGYAKTNGHSYDMEDGSVFLFTSESVGEGHPDKMCDQISDAILDAHLNQDPDAKVACETITKTGMVLLCGEITSKANVDYQKVVRETVKHIGYDDSSKGFDYKTCSVMLALDQQSPNIAAGVHENRNDEEVGAGDQGLMFGYATDETEECMPLTVVLAHKLNQKIAELRRNGEFWWARPDSKTQVTCEYVFAGGATVPQRVHTVVVSLQHSEKITLETLRDEIREKVIKEVIPAQYLDERTVIHINPCGLFIIGGPQSDAGLTGRKIIVDTYGGWGAHGGGAFSGKDFTKVDRSAAYAARWVAKSLVKAGLCRRCMVQVAYAIGVAEPLSITVFDYGTSHKTQQQLLSIVQKNFDLRPGKIVKELNLRAPIYQRTSTYGHFGRAGFPWESPKPLIVE
Summary
Description
Catalyzes the formation of S-adenosylmethionine from methionine and ATP.
Catalyzes the formation of S-adenosylmethionine from methionine and ATP. The reaction comprises two steps that are both catalyzed by the same enzyme: formation of S-adenosylmethionine (AdoMet) and triphosphate, and subsequent hydrolysis of the triphosphate.
Catalyzes the formation of S-adenosylmethionine (AdoMet) from methionine and ATP. The overall synthetic reaction is composed of two sequential steps, AdoMet formation and the subsequent tripolyphosphate hydrolysis which occurs prior to release of AdoMet from the enzyme.
Catalyzes the formation of S-adenosylmethionine from methionine and ATP. The reaction comprises two steps that are both catalyzed by the same enzyme: formation of S-adenosylmethionine (AdoMet) and triphosphate, and subsequent hydrolysis of the triphosphate.
Catalyzes the formation of S-adenosylmethionine (AdoMet) from methionine and ATP. The overall synthetic reaction is composed of two sequential steps, AdoMet formation and the subsequent tripolyphosphate hydrolysis which occurs prior to release of AdoMet from the enzyme.
Catalytic Activity
ATP + H2O + L-methionine = diphosphate + phosphate + S-adenosyl-L-methionine
Cofactor
K(+)
Mg(2+)
Mg(2+)
Subunit
Homotetramer; dimer of dimers.
Similarity
Belongs to the AdoMet synthase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Magnesium
Metal-binding
Nucleotide-binding
One-carbon metabolism
Potassium
Reference proteome
Transferase
Feature
chain S-adenosylmethionine synthase
splice variant In isoform 4.
splice variant In isoform 4.
Uniprot
Q1HQB5
A0A2H1WQI2
H9JSK4
A0A3S2L3K0
A0A0T6AV40
N6T4Q1
+ More
Q0IFM6 A0A2M3YX87 A0A2M4BPV6 W5J5M5 Q7PTN3 A0A2M4A0J8 A0A084VPR9 T1DI67 T1E8C0 A0A1W4X5D8 A0A1Q3F3Q3 A0A158NRC9 A0A2M3ZZ46 B0W244 A0A182RGA4 A0A1Q3F3T2 A0A1Q3F3Q7 K7J547 A0A0K8TNX2 V9ICM2 A0A0C9QJ66 T1PCY3 A0A034VY08 A0A1Y1MTA1 V5GQX9 A0A1S3D847 A0A1Q3FWJ2 A0A2J7RI80 A0A1W4X411 A0A1I8PNE2 A0A023ESR9 A0A0J9QTU8 A0A0Q5W8R3 P40320 A0A1W4V3Z4 B4P1V0 A0A2H8TWK6 A0A1S4H166 A0A0A1WZ25 J9JXG1 A0A1I8NA59 A0A1L8DZP2 B3MLT7 A0A1J1HV82 B4N0M1 A0A0P6IPP1 E0VI81 A0A3B0JUP6 A0A0J9QSU3 B3N770 P40320-3 A4UZW2 B4GQT3 A0A0R3NRQ1 A0A0R1DIG4 A0A1S3D9I4 J9JWP7 A0A1B6H484 A0A0A9VPX0 A0A1W4UQK6 B4ICR7 A0A1Y1MT92 A0A0L0BLR5 A0A0N8NZ13 A0A1I8PNH7 A0A069DYW6 T1I729 A0A1J1HV75 A0A023F8X8 A0A1B6L7Z3 W8BP23 A0A1A9ZJ02 B4MDB4 R4WRS0 A0A3B0K2G9 A0A0Q9WR93 Q29NR2 A0A1A9V8X4 D3TPX3 A0A1L8DZK7 A0A1A9W0H9 A0A2R7W5M8 A0A0A9VWE3 A0A0Q9WV10 R4FQA5 A0A0H4IRI4 A0A336MJF9 A0A1B6I9T0 A0A067R1V0 A0A1B6J7T8 A0A0P4WB30 A0A226EU99 A0A323TLE5 A0A3R7MAV7
Q0IFM6 A0A2M3YX87 A0A2M4BPV6 W5J5M5 Q7PTN3 A0A2M4A0J8 A0A084VPR9 T1DI67 T1E8C0 A0A1W4X5D8 A0A1Q3F3Q3 A0A158NRC9 A0A2M3ZZ46 B0W244 A0A182RGA4 A0A1Q3F3T2 A0A1Q3F3Q7 K7J547 A0A0K8TNX2 V9ICM2 A0A0C9QJ66 T1PCY3 A0A034VY08 A0A1Y1MTA1 V5GQX9 A0A1S3D847 A0A1Q3FWJ2 A0A2J7RI80 A0A1W4X411 A0A1I8PNE2 A0A023ESR9 A0A0J9QTU8 A0A0Q5W8R3 P40320 A0A1W4V3Z4 B4P1V0 A0A2H8TWK6 A0A1S4H166 A0A0A1WZ25 J9JXG1 A0A1I8NA59 A0A1L8DZP2 B3MLT7 A0A1J1HV82 B4N0M1 A0A0P6IPP1 E0VI81 A0A3B0JUP6 A0A0J9QSU3 B3N770 P40320-3 A4UZW2 B4GQT3 A0A0R3NRQ1 A0A0R1DIG4 A0A1S3D9I4 J9JWP7 A0A1B6H484 A0A0A9VPX0 A0A1W4UQK6 B4ICR7 A0A1Y1MT92 A0A0L0BLR5 A0A0N8NZ13 A0A1I8PNH7 A0A069DYW6 T1I729 A0A1J1HV75 A0A023F8X8 A0A1B6L7Z3 W8BP23 A0A1A9ZJ02 B4MDB4 R4WRS0 A0A3B0K2G9 A0A0Q9WR93 Q29NR2 A0A1A9V8X4 D3TPX3 A0A1L8DZK7 A0A1A9W0H9 A0A2R7W5M8 A0A0A9VWE3 A0A0Q9WV10 R4FQA5 A0A0H4IRI4 A0A336MJF9 A0A1B6I9T0 A0A067R1V0 A0A1B6J7T8 A0A0P4WB30 A0A226EU99 A0A323TLE5 A0A3R7MAV7
EC Number
2.5.1.6
Pubmed
19121390
23537049
17510324
20920257
23761445
12364791
+ More
14747013 17210077 24438588 24330624 21347285 20075255 26369729 25315136 25348373 28004739 24945155 26483478 22936249 17994087 18057021 8150093 10731132 12537572 12537569 17550304 25830018 20566863 12537568 12537573 12537574 16110336 17569856 17569867 15632085 23185243 25401762 26823975 26108605 26334808 25474469 24495485 23691247 20353571 24845553
14747013 17210077 24438588 24330624 21347285 20075255 26369729 25315136 25348373 28004739 24945155 26483478 22936249 17994087 18057021 8150093 10731132 12537572 12537569 17550304 25830018 20566863 12537568 12537573 12537574 16110336 17569856 17569867 15632085 23185243 25401762 26823975 26108605 26334808 25474469 24495485 23691247 20353571 24845553
EMBL
DQ443137
ABF51226.1
ODYU01010301
SOQ55313.1
BABH01028031
RSAL01000185
+ More
RVE44816.1 LJIG01022734 KRT78988.1 APGK01052480 KB741213 KB631625 ENN72583.1 ERL84820.1 CH477306 EAT44117.1 GGFM01000138 MBW20889.1 GGFJ01005974 MBW55115.1 ADMH02002093 ETN59291.1 AAAB01008797 EAA03629.2 GGFK01000950 MBW34271.1 ATLV01015034 KE524999 KFB39963.1 GALA01001111 JAA93741.1 GAMD01002903 JAA98687.1 GFDL01012872 JAV22173.1 ADTU01023878 ADTU01023879 GGFK01000409 MBW33730.1 DS231825 DS232411 EDS28172.1 EDS41283.1 GFDL01012866 JAV22179.1 GFDL01012849 JAV22196.1 AAZX01000337 GDAI01001772 JAI15831.1 JR038775 AEY58407.1 GBYB01014768 JAG84535.1 KA646524 AFP61153.1 GAKP01012223 GAKP01012222 JAC46730.1 GEZM01021619 JAV88902.1 GALX01004479 JAB63987.1 GFDL01003120 JAV31925.1 NEVH01003505 PNF40533.1 JXUM01092894 JXUM01104861 JXUM01104862 GAPW01001762 KQ564921 KQ564018 JAC11836.1 KXJ71573.1 KXJ72925.1 CM002910 KMY87234.1 CH954177 KQS69900.1 X77392 AE014134 AY051918 CM000157 EDW87086.2 GFXV01006870 MBW18675.1 GBXI01010185 JAD04107.1 ABLF02028542 GFDF01002187 JAV11897.1 CH902620 EDV30808.2 KPU73042.1 CVRI01000021 CRK91984.1 CH963920 EDW77634.2 GDIQ01002231 JAN92506.1 DS235184 EEB13087.1 OUUW01000004 SPP79200.1 KMY87232.1 EDV57177.1 AAN10504.1 CH479187 EDW39955.1 CH379059 KRT03689.1 KRJ97015.1 ABLF02027607 GECZ01000264 JAS69505.1 GBHO01045083 GBHO01045082 GBHO01045079 GBRD01002445 GDHC01000816 JAF98520.1 JAF98521.1 JAF98524.1 JAG63376.1 JAQ17813.1 CH480829 EDW45343.1 GEZM01021618 JAV88903.1 JRES01001684 KNC20967.1 KPU73044.1 GBGD01001675 JAC87214.1 ACPB03024900 ACPB03024901 CRK91983.1 GBBI01000820 JAC17892.1 GEBQ01020159 JAT19818.1 GAMC01015102 GAMC01015101 JAB91454.1 CH940661 EDW71175.1 KRF85501.1 AK417397 BAN20612.1 SPP79201.1 KRF98714.1 EAL34156.2 KRT03690.1 EZ423475 ADD19751.1 GFDF01002174 JAV11910.1 KK854356 PTY15036.1 GBHO01045081 GBHO01045080 GBHO01045078 GBRD01002447 JAF98522.1 JAF98523.1 JAF98525.1 JAG63374.1 KRF85505.1 GAHY01000514 JAA76996.1 KP689344 AKO63326.1 UFQT01001434 SSX30542.1 GECU01024039 JAS83667.1 KK852809 KDR15976.1 GECU01012460 JAS95246.1 GDRN01057346 JAI65702.1 LNIX01000002 OXA60196.1 MDVM02000056 PYZ99334.1 QCYY01001486 ROT77738.1
RVE44816.1 LJIG01022734 KRT78988.1 APGK01052480 KB741213 KB631625 ENN72583.1 ERL84820.1 CH477306 EAT44117.1 GGFM01000138 MBW20889.1 GGFJ01005974 MBW55115.1 ADMH02002093 ETN59291.1 AAAB01008797 EAA03629.2 GGFK01000950 MBW34271.1 ATLV01015034 KE524999 KFB39963.1 GALA01001111 JAA93741.1 GAMD01002903 JAA98687.1 GFDL01012872 JAV22173.1 ADTU01023878 ADTU01023879 GGFK01000409 MBW33730.1 DS231825 DS232411 EDS28172.1 EDS41283.1 GFDL01012866 JAV22179.1 GFDL01012849 JAV22196.1 AAZX01000337 GDAI01001772 JAI15831.1 JR038775 AEY58407.1 GBYB01014768 JAG84535.1 KA646524 AFP61153.1 GAKP01012223 GAKP01012222 JAC46730.1 GEZM01021619 JAV88902.1 GALX01004479 JAB63987.1 GFDL01003120 JAV31925.1 NEVH01003505 PNF40533.1 JXUM01092894 JXUM01104861 JXUM01104862 GAPW01001762 KQ564921 KQ564018 JAC11836.1 KXJ71573.1 KXJ72925.1 CM002910 KMY87234.1 CH954177 KQS69900.1 X77392 AE014134 AY051918 CM000157 EDW87086.2 GFXV01006870 MBW18675.1 GBXI01010185 JAD04107.1 ABLF02028542 GFDF01002187 JAV11897.1 CH902620 EDV30808.2 KPU73042.1 CVRI01000021 CRK91984.1 CH963920 EDW77634.2 GDIQ01002231 JAN92506.1 DS235184 EEB13087.1 OUUW01000004 SPP79200.1 KMY87232.1 EDV57177.1 AAN10504.1 CH479187 EDW39955.1 CH379059 KRT03689.1 KRJ97015.1 ABLF02027607 GECZ01000264 JAS69505.1 GBHO01045083 GBHO01045082 GBHO01045079 GBRD01002445 GDHC01000816 JAF98520.1 JAF98521.1 JAF98524.1 JAG63376.1 JAQ17813.1 CH480829 EDW45343.1 GEZM01021618 JAV88903.1 JRES01001684 KNC20967.1 KPU73044.1 GBGD01001675 JAC87214.1 ACPB03024900 ACPB03024901 CRK91983.1 GBBI01000820 JAC17892.1 GEBQ01020159 JAT19818.1 GAMC01015102 GAMC01015101 JAB91454.1 CH940661 EDW71175.1 KRF85501.1 AK417397 BAN20612.1 SPP79201.1 KRF98714.1 EAL34156.2 KRT03690.1 EZ423475 ADD19751.1 GFDF01002174 JAV11910.1 KK854356 PTY15036.1 GBHO01045081 GBHO01045080 GBHO01045078 GBRD01002447 JAF98522.1 JAF98523.1 JAF98525.1 JAG63374.1 KRF85505.1 GAHY01000514 JAA76996.1 KP689344 AKO63326.1 UFQT01001434 SSX30542.1 GECU01024039 JAS83667.1 KK852809 KDR15976.1 GECU01012460 JAS95246.1 GDRN01057346 JAI65702.1 LNIX01000002 OXA60196.1 MDVM02000056 PYZ99334.1 QCYY01001486 ROT77738.1
Proteomes
UP000005204
UP000283053
UP000019118
UP000030742
UP000008820
UP000000673
+ More
UP000007062 UP000030765 UP000192223 UP000005205 UP000002320 UP000075900 UP000002358 UP000095301 UP000079169 UP000235965 UP000095300 UP000069940 UP000249989 UP000008711 UP000000803 UP000192221 UP000002282 UP000007819 UP000007801 UP000183832 UP000007798 UP000009046 UP000268350 UP000008744 UP000001819 UP000001292 UP000037069 UP000015103 UP000092445 UP000008792 UP000078200 UP000091820 UP000027135 UP000198287 UP000247427 UP000283509
UP000007062 UP000030765 UP000192223 UP000005205 UP000002320 UP000075900 UP000002358 UP000095301 UP000079169 UP000235965 UP000095300 UP000069940 UP000249989 UP000008711 UP000000803 UP000192221 UP000002282 UP000007819 UP000007801 UP000183832 UP000007798 UP000009046 UP000268350 UP000008744 UP000001819 UP000001292 UP000037069 UP000015103 UP000092445 UP000008792 UP000078200 UP000091820 UP000027135 UP000198287 UP000247427 UP000283509
Interpro
SUPFAM
SSF55973
SSF55973
ProteinModelPortal
Q1HQB5
A0A2H1WQI2
H9JSK4
A0A3S2L3K0
A0A0T6AV40
N6T4Q1
+ More
Q0IFM6 A0A2M3YX87 A0A2M4BPV6 W5J5M5 Q7PTN3 A0A2M4A0J8 A0A084VPR9 T1DI67 T1E8C0 A0A1W4X5D8 A0A1Q3F3Q3 A0A158NRC9 A0A2M3ZZ46 B0W244 A0A182RGA4 A0A1Q3F3T2 A0A1Q3F3Q7 K7J547 A0A0K8TNX2 V9ICM2 A0A0C9QJ66 T1PCY3 A0A034VY08 A0A1Y1MTA1 V5GQX9 A0A1S3D847 A0A1Q3FWJ2 A0A2J7RI80 A0A1W4X411 A0A1I8PNE2 A0A023ESR9 A0A0J9QTU8 A0A0Q5W8R3 P40320 A0A1W4V3Z4 B4P1V0 A0A2H8TWK6 A0A1S4H166 A0A0A1WZ25 J9JXG1 A0A1I8NA59 A0A1L8DZP2 B3MLT7 A0A1J1HV82 B4N0M1 A0A0P6IPP1 E0VI81 A0A3B0JUP6 A0A0J9QSU3 B3N770 P40320-3 A4UZW2 B4GQT3 A0A0R3NRQ1 A0A0R1DIG4 A0A1S3D9I4 J9JWP7 A0A1B6H484 A0A0A9VPX0 A0A1W4UQK6 B4ICR7 A0A1Y1MT92 A0A0L0BLR5 A0A0N8NZ13 A0A1I8PNH7 A0A069DYW6 T1I729 A0A1J1HV75 A0A023F8X8 A0A1B6L7Z3 W8BP23 A0A1A9ZJ02 B4MDB4 R4WRS0 A0A3B0K2G9 A0A0Q9WR93 Q29NR2 A0A1A9V8X4 D3TPX3 A0A1L8DZK7 A0A1A9W0H9 A0A2R7W5M8 A0A0A9VWE3 A0A0Q9WV10 R4FQA5 A0A0H4IRI4 A0A336MJF9 A0A1B6I9T0 A0A067R1V0 A0A1B6J7T8 A0A0P4WB30 A0A226EU99 A0A323TLE5 A0A3R7MAV7
Q0IFM6 A0A2M3YX87 A0A2M4BPV6 W5J5M5 Q7PTN3 A0A2M4A0J8 A0A084VPR9 T1DI67 T1E8C0 A0A1W4X5D8 A0A1Q3F3Q3 A0A158NRC9 A0A2M3ZZ46 B0W244 A0A182RGA4 A0A1Q3F3T2 A0A1Q3F3Q7 K7J547 A0A0K8TNX2 V9ICM2 A0A0C9QJ66 T1PCY3 A0A034VY08 A0A1Y1MTA1 V5GQX9 A0A1S3D847 A0A1Q3FWJ2 A0A2J7RI80 A0A1W4X411 A0A1I8PNE2 A0A023ESR9 A0A0J9QTU8 A0A0Q5W8R3 P40320 A0A1W4V3Z4 B4P1V0 A0A2H8TWK6 A0A1S4H166 A0A0A1WZ25 J9JXG1 A0A1I8NA59 A0A1L8DZP2 B3MLT7 A0A1J1HV82 B4N0M1 A0A0P6IPP1 E0VI81 A0A3B0JUP6 A0A0J9QSU3 B3N770 P40320-3 A4UZW2 B4GQT3 A0A0R3NRQ1 A0A0R1DIG4 A0A1S3D9I4 J9JWP7 A0A1B6H484 A0A0A9VPX0 A0A1W4UQK6 B4ICR7 A0A1Y1MT92 A0A0L0BLR5 A0A0N8NZ13 A0A1I8PNH7 A0A069DYW6 T1I729 A0A1J1HV75 A0A023F8X8 A0A1B6L7Z3 W8BP23 A0A1A9ZJ02 B4MDB4 R4WRS0 A0A3B0K2G9 A0A0Q9WR93 Q29NR2 A0A1A9V8X4 D3TPX3 A0A1L8DZK7 A0A1A9W0H9 A0A2R7W5M8 A0A0A9VWE3 A0A0Q9WV10 R4FQA5 A0A0H4IRI4 A0A336MJF9 A0A1B6I9T0 A0A067R1V0 A0A1B6J7T8 A0A0P4WB30 A0A226EU99 A0A323TLE5 A0A3R7MAV7
PDB
6FWB
E-value=1.4778e-169,
Score=1530
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
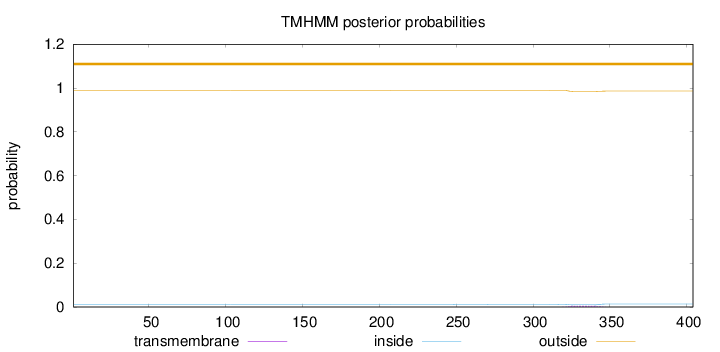
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11316
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01111
outside
1 - 404
Population Genetic Test Statistics
Pi
268.608058
Theta
214.257659
Tajima's D
1.034085
CLR
0.225878
CSRT
0.665366731663417
Interpretation
Uncertain
