Gene
KWMTBOMO05089 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012451
Annotation
SCP-related_protein_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.996
Sequence
CDS
ATGCAGTCGTTCCTTATATTTGTTGTCGCACTCGCGTGCTTTCAATCTATTGATTGTCGGAAGTTACAGCCATTATCGTGCGACGATATACGACAGTTCGTCAACGGTCACAATTTGCGGCGTGAACAAATAGCCAAGGGTGAAATATCTGGACAGCCGGCAGCGACCCAAATGAAATACATGATTTGGGATAAGGAGTTAGCTGCGAAAGCTGCAAAGTGGGCTTCGTCGGACAATGATTTCCATAATCCAGACAGGAGCATAGCATCCGAAAGGTTTTCAACGGGCGAGAACTTGTACTGGTATATGACGAGCAATAAAAATCACAAGATCAACCCAGATTCCGCCCTCGAATCTTGGTTCAATGAACATGAAAACTACAAATTTGCGCCCTTAAAATCGAGCGACTTTCAAAAAACTGGGAAAAAGCAGATCGCCCACTACACTCAGATGGTGTGGTCTGATAGTGATCGCGTTGGCTGCGCTATCGGCACCTCGCGGACTGCACAGATGAAATCATTCTTCGTTGTGTGCAACTATGGACCTGCTGGGAACTATCTAGGCAATATCCCATACAAATCCGGAGAGCCGAGCAATCGTCTCGTCTGCGGCGCCGGCGACTGCAGTAAACCTTATGGAGATCAATGCTAA
Protein
MQSFLIFVVALACFQSIDCRKLQPLSCDDIRQFVNGHNLRREQIAKGEISGQPAATQMKYMIWDKELAAKAAKWASSDNDFHNPDRSIASERFSTGENLYWYMTSNKNHKINPDSALESWFNEHENYKFAPLKSSDFQKTGKKQIAHYTQMVWSDSDRVGCAIGTSRTAQMKSFFVVCNYGPAGNYLGNIPYKSGEPSNRLVCGAGDCSKPYGDQC
Summary
Similarity
Belongs to the CRISP family.
Uniprot
Q152R4
A0A3S2NUE2
A0A194RQ07
A0A194PE84
A0A194RQ92
A0A2H1VKX9
+ More
A0A194PDK8 S4NY11 A0A212EIP5 A0A3S2TFT4 A0A3S2LUT5 A0A194PE94 A0A3S2P741 A0A212F956 E2ABL4 K7IUB0 A0A0A9VVM5 A0A146LDR5 A0A1B6LJ27 A0A1I9WL95 A0A232FDU7 D6W9F0 K7ITU2 A0A1B0CS01 W8AJX1 A0A0A1WD73 A0A0K8UYG2 A0A1I9WL94 A0A0R3NGL4 B4G5N0 A0A0N8BUM3 A0A034W1K3 A0A0P5CGW3 A0A0K8U2B7 E9G1H2 A0A0P5X1V0 A0A0P5J1B6 A0A0A9XWZ4 A0A0P5GLS6 A0A0P5LJ85 A0A0P5NP70 A0A146LE22 A0A0P6GS87 A0A0P5HJA8 A0A0P5J2Y1 A0A0P5XKW7 A0A0P6GA61 A0A0P4YXM5 A0A0P5YDZ3 A0A3B0KHI6 A0A0P6BDU1 Q299S2 A0A0P5DWN1 B3LX49 A0A1A9XSB9 A0A1B0DJF6 A0A0P6I300 A0A146MDA6 B4NG58 B3P175 A0A1I8P6F4 A0A336M4G4 B4JEU6 A0A0A9WQ81 B4PPX8 Q9VFY2 B4R1V3 A0A0C9QJT4 E9FTJ2 A0A0P4YUC3 A0A336M7K6 A0A067XK13 A0A310SR82 B4HGA3 A0A1W4UJ18 B4K8G3 A0A1A9UNT7 A0A2R7W7S1 A0A232F7Q8 A0A1I8NCD4 A0A1A9Z725 K7ITU1 A0A336MFL5 A0A0P5WBI8 N6TQ74 A0A336N4I2 U4TY95 B4M6K7 A0A0M4ER11 A0A0L0BX64 T1PA51 A0A224XJN9 A0A026WK75 A0A2R7VP63 A0A3L8DBV3 A0A1L8DRA4 A0A2P8YFD4 A0A085LW55 A0A1L8D8Q4
A0A194PDK8 S4NY11 A0A212EIP5 A0A3S2TFT4 A0A3S2LUT5 A0A194PE94 A0A3S2P741 A0A212F956 E2ABL4 K7IUB0 A0A0A9VVM5 A0A146LDR5 A0A1B6LJ27 A0A1I9WL95 A0A232FDU7 D6W9F0 K7ITU2 A0A1B0CS01 W8AJX1 A0A0A1WD73 A0A0K8UYG2 A0A1I9WL94 A0A0R3NGL4 B4G5N0 A0A0N8BUM3 A0A034W1K3 A0A0P5CGW3 A0A0K8U2B7 E9G1H2 A0A0P5X1V0 A0A0P5J1B6 A0A0A9XWZ4 A0A0P5GLS6 A0A0P5LJ85 A0A0P5NP70 A0A146LE22 A0A0P6GS87 A0A0P5HJA8 A0A0P5J2Y1 A0A0P5XKW7 A0A0P6GA61 A0A0P4YXM5 A0A0P5YDZ3 A0A3B0KHI6 A0A0P6BDU1 Q299S2 A0A0P5DWN1 B3LX49 A0A1A9XSB9 A0A1B0DJF6 A0A0P6I300 A0A146MDA6 B4NG58 B3P175 A0A1I8P6F4 A0A336M4G4 B4JEU6 A0A0A9WQ81 B4PPX8 Q9VFY2 B4R1V3 A0A0C9QJT4 E9FTJ2 A0A0P4YUC3 A0A336M7K6 A0A067XK13 A0A310SR82 B4HGA3 A0A1W4UJ18 B4K8G3 A0A1A9UNT7 A0A2R7W7S1 A0A232F7Q8 A0A1I8NCD4 A0A1A9Z725 K7ITU1 A0A336MFL5 A0A0P5WBI8 N6TQ74 A0A336N4I2 U4TY95 B4M6K7 A0A0M4ER11 A0A0L0BX64 T1PA51 A0A224XJN9 A0A026WK75 A0A2R7VP63 A0A3L8DBV3 A0A1L8DRA4 A0A2P8YFD4 A0A085LW55 A0A1L8D8Q4
Pubmed
19121390
26354079
23622113
22118469
20798317
20075255
+ More
25401762 26823975 27538518 28648823 18362917 19820115 24495485 25830018 15632085 17994087 25348373 21292972 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 23537049 26108605 24508170 30249741 29403074 24929829
25401762 26823975 27538518 28648823 18362917 19820115 24495485 25830018 15632085 17994087 25348373 21292972 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 23537049 26108605 24508170 30249741 29403074 24929829
EMBL
BABH01028031
DQ652543
ABG25817.1
RSAL01000185
RVE44819.1
KQ459896
+ More
KPJ19489.1 KQ459606 KPI91338.1 KPJ19490.1 ODYU01003124 SOQ41490.1 KPI91337.1 GAIX01010446 JAA82114.1 AGBW02014611 OWR41341.1 RVE44821.1 RVE44822.1 KPI91348.1 RSAL01000257 RVE43403.1 AGBW02009645 OWR50261.1 GL438297 EFN69185.1 AAZX01010653 GBHO01043870 JAF99733.1 GDHC01012176 JAQ06453.1 GEBQ01016280 JAT23697.1 KU932279 APA33915.1 NNAY01000357 OXU28931.1 KQ971312 EEZ98167.1 AJWK01025522 GAMC01021567 GAMC01021566 JAB84988.1 GBXI01017353 JAC96938.1 GDHF01020668 JAI31646.1 KU932278 APA33914.1 CM000070 KRT00193.1 CH479179 EDW24896.1 GDIQ01143275 JAL08451.1 GAKP01010937 JAC48015.1 GDIP01174440 JAJ48962.1 GDHF01031663 JAI20651.1 GL732529 EFX86816.1 GDIP01078608 JAM25107.1 GDIQ01205901 JAK45824.1 GBHO01020216 GBRD01000114 GDHC01011013 JAG23388.1 JAG65707.1 JAQ07616.1 GDIQ01245309 JAK06416.1 GDIQ01168823 JAK82902.1 GDIQ01139639 JAL12087.1 GDHC01012115 JAQ06514.1 GDIQ01039674 JAN55063.1 GDIQ01227213 JAK24512.1 GDIQ01207304 JAK44421.1 GDIP01105949 GDIP01070856 LRGB01003163 JAM32859.1 KZS03911.1 GDIQ01036331 JAN58406.1 GDIP01222192 JAJ01210.1 GDIP01147905 GDIP01105948 GDIP01059169 JAM44546.1 OUUW01000013 SPP87900.1 GDIP01016581 JAM87134.1 EAL27629.2 GDIP01150546 JAJ72856.1 CH902617 EDV41649.2 AJVK01015784 GDIQ01011264 JAN83473.1 GDHC01001903 JAQ16726.1 CH964251 EDW83275.1 CH954181 EDV49264.1 UFQT01000512 SSX24860.1 CH916369 EDV93227.1 GBHO01034976 JAG08628.1 CM000160 EDW97205.1 AE014297 AY051901 AAF54913.2 AAK93325.1 CM000364 EDX13105.1 GBYB01000862 JAG70629.1 GL733862 GL732524 EFX62063.1 EFX89372.1 GDIP01223485 JAI99916.1 SSX24859.1 JX399878 AGI44426.1 KQ761389 OAD57679.1 CH480815 EDW42351.1 CH933806 EDW14362.1 KK854426 PTY15763.1 NNAY01000705 OXU26884.1 UFQT01000511 SSX24858.1 GDIP01088405 JAM15310.1 APGK01025243 APGK01048302 KB741104 KB740579 ENN73843.1 ENN80178.1 UFQT01002378 SSX33338.1 KB631831 ERL86594.1 CH940652 EDW59283.1 CP012526 ALC47538.1 JRES01001300 KNC23824.1 KA644818 AFP59447.1 GFTR01003720 JAW12706.1 KK107167 EZA56452.1 KK854013 PTY09327.1 QOIP01000010 RLU17977.1 GFDF01005219 JAV08865.1 PYGN01000641 PSN42953.1 KL363276 KL367557 KFD49201.1 KFD64323.1 GFDF01011252 JAV02832.1
KPJ19489.1 KQ459606 KPI91338.1 KPJ19490.1 ODYU01003124 SOQ41490.1 KPI91337.1 GAIX01010446 JAA82114.1 AGBW02014611 OWR41341.1 RVE44821.1 RVE44822.1 KPI91348.1 RSAL01000257 RVE43403.1 AGBW02009645 OWR50261.1 GL438297 EFN69185.1 AAZX01010653 GBHO01043870 JAF99733.1 GDHC01012176 JAQ06453.1 GEBQ01016280 JAT23697.1 KU932279 APA33915.1 NNAY01000357 OXU28931.1 KQ971312 EEZ98167.1 AJWK01025522 GAMC01021567 GAMC01021566 JAB84988.1 GBXI01017353 JAC96938.1 GDHF01020668 JAI31646.1 KU932278 APA33914.1 CM000070 KRT00193.1 CH479179 EDW24896.1 GDIQ01143275 JAL08451.1 GAKP01010937 JAC48015.1 GDIP01174440 JAJ48962.1 GDHF01031663 JAI20651.1 GL732529 EFX86816.1 GDIP01078608 JAM25107.1 GDIQ01205901 JAK45824.1 GBHO01020216 GBRD01000114 GDHC01011013 JAG23388.1 JAG65707.1 JAQ07616.1 GDIQ01245309 JAK06416.1 GDIQ01168823 JAK82902.1 GDIQ01139639 JAL12087.1 GDHC01012115 JAQ06514.1 GDIQ01039674 JAN55063.1 GDIQ01227213 JAK24512.1 GDIQ01207304 JAK44421.1 GDIP01105949 GDIP01070856 LRGB01003163 JAM32859.1 KZS03911.1 GDIQ01036331 JAN58406.1 GDIP01222192 JAJ01210.1 GDIP01147905 GDIP01105948 GDIP01059169 JAM44546.1 OUUW01000013 SPP87900.1 GDIP01016581 JAM87134.1 EAL27629.2 GDIP01150546 JAJ72856.1 CH902617 EDV41649.2 AJVK01015784 GDIQ01011264 JAN83473.1 GDHC01001903 JAQ16726.1 CH964251 EDW83275.1 CH954181 EDV49264.1 UFQT01000512 SSX24860.1 CH916369 EDV93227.1 GBHO01034976 JAG08628.1 CM000160 EDW97205.1 AE014297 AY051901 AAF54913.2 AAK93325.1 CM000364 EDX13105.1 GBYB01000862 JAG70629.1 GL733862 GL732524 EFX62063.1 EFX89372.1 GDIP01223485 JAI99916.1 SSX24859.1 JX399878 AGI44426.1 KQ761389 OAD57679.1 CH480815 EDW42351.1 CH933806 EDW14362.1 KK854426 PTY15763.1 NNAY01000705 OXU26884.1 UFQT01000511 SSX24858.1 GDIP01088405 JAM15310.1 APGK01025243 APGK01048302 KB741104 KB740579 ENN73843.1 ENN80178.1 UFQT01002378 SSX33338.1 KB631831 ERL86594.1 CH940652 EDW59283.1 CP012526 ALC47538.1 JRES01001300 KNC23824.1 KA644818 AFP59447.1 GFTR01003720 JAW12706.1 KK107167 EZA56452.1 KK854013 PTY09327.1 QOIP01000010 RLU17977.1 GFDF01005219 JAV08865.1 PYGN01000641 PSN42953.1 KL363276 KL367557 KFD49201.1 KFD64323.1 GFDF01011252 JAV02832.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000000311
+ More
UP000002358 UP000215335 UP000007266 UP000092461 UP000001819 UP000008744 UP000000305 UP000076858 UP000268350 UP000007801 UP000092443 UP000092462 UP000007798 UP000008711 UP000095300 UP000001070 UP000002282 UP000000803 UP000000304 UP000001292 UP000192221 UP000009192 UP000078200 UP000095301 UP000092445 UP000019118 UP000030742 UP000008792 UP000092553 UP000037069 UP000053097 UP000279307 UP000245037
UP000002358 UP000215335 UP000007266 UP000092461 UP000001819 UP000008744 UP000000305 UP000076858 UP000268350 UP000007801 UP000092443 UP000092462 UP000007798 UP000008711 UP000095300 UP000001070 UP000002282 UP000000803 UP000000304 UP000001292 UP000192221 UP000009192 UP000078200 UP000095301 UP000092445 UP000019118 UP000030742 UP000008792 UP000092553 UP000037069 UP000053097 UP000279307 UP000245037
Interpro
SUPFAM
SSF55797
SSF55797
Gene 3D
ProteinModelPortal
Q152R4
A0A3S2NUE2
A0A194RQ07
A0A194PE84
A0A194RQ92
A0A2H1VKX9
+ More
A0A194PDK8 S4NY11 A0A212EIP5 A0A3S2TFT4 A0A3S2LUT5 A0A194PE94 A0A3S2P741 A0A212F956 E2ABL4 K7IUB0 A0A0A9VVM5 A0A146LDR5 A0A1B6LJ27 A0A1I9WL95 A0A232FDU7 D6W9F0 K7ITU2 A0A1B0CS01 W8AJX1 A0A0A1WD73 A0A0K8UYG2 A0A1I9WL94 A0A0R3NGL4 B4G5N0 A0A0N8BUM3 A0A034W1K3 A0A0P5CGW3 A0A0K8U2B7 E9G1H2 A0A0P5X1V0 A0A0P5J1B6 A0A0A9XWZ4 A0A0P5GLS6 A0A0P5LJ85 A0A0P5NP70 A0A146LE22 A0A0P6GS87 A0A0P5HJA8 A0A0P5J2Y1 A0A0P5XKW7 A0A0P6GA61 A0A0P4YXM5 A0A0P5YDZ3 A0A3B0KHI6 A0A0P6BDU1 Q299S2 A0A0P5DWN1 B3LX49 A0A1A9XSB9 A0A1B0DJF6 A0A0P6I300 A0A146MDA6 B4NG58 B3P175 A0A1I8P6F4 A0A336M4G4 B4JEU6 A0A0A9WQ81 B4PPX8 Q9VFY2 B4R1V3 A0A0C9QJT4 E9FTJ2 A0A0P4YUC3 A0A336M7K6 A0A067XK13 A0A310SR82 B4HGA3 A0A1W4UJ18 B4K8G3 A0A1A9UNT7 A0A2R7W7S1 A0A232F7Q8 A0A1I8NCD4 A0A1A9Z725 K7ITU1 A0A336MFL5 A0A0P5WBI8 N6TQ74 A0A336N4I2 U4TY95 B4M6K7 A0A0M4ER11 A0A0L0BX64 T1PA51 A0A224XJN9 A0A026WK75 A0A2R7VP63 A0A3L8DBV3 A0A1L8DRA4 A0A2P8YFD4 A0A085LW55 A0A1L8D8Q4
A0A194PDK8 S4NY11 A0A212EIP5 A0A3S2TFT4 A0A3S2LUT5 A0A194PE94 A0A3S2P741 A0A212F956 E2ABL4 K7IUB0 A0A0A9VVM5 A0A146LDR5 A0A1B6LJ27 A0A1I9WL95 A0A232FDU7 D6W9F0 K7ITU2 A0A1B0CS01 W8AJX1 A0A0A1WD73 A0A0K8UYG2 A0A1I9WL94 A0A0R3NGL4 B4G5N0 A0A0N8BUM3 A0A034W1K3 A0A0P5CGW3 A0A0K8U2B7 E9G1H2 A0A0P5X1V0 A0A0P5J1B6 A0A0A9XWZ4 A0A0P5GLS6 A0A0P5LJ85 A0A0P5NP70 A0A146LE22 A0A0P6GS87 A0A0P5HJA8 A0A0P5J2Y1 A0A0P5XKW7 A0A0P6GA61 A0A0P4YXM5 A0A0P5YDZ3 A0A3B0KHI6 A0A0P6BDU1 Q299S2 A0A0P5DWN1 B3LX49 A0A1A9XSB9 A0A1B0DJF6 A0A0P6I300 A0A146MDA6 B4NG58 B3P175 A0A1I8P6F4 A0A336M4G4 B4JEU6 A0A0A9WQ81 B4PPX8 Q9VFY2 B4R1V3 A0A0C9QJT4 E9FTJ2 A0A0P4YUC3 A0A336M7K6 A0A067XK13 A0A310SR82 B4HGA3 A0A1W4UJ18 B4K8G3 A0A1A9UNT7 A0A2R7W7S1 A0A232F7Q8 A0A1I8NCD4 A0A1A9Z725 K7ITU1 A0A336MFL5 A0A0P5WBI8 N6TQ74 A0A336N4I2 U4TY95 B4M6K7 A0A0M4ER11 A0A0L0BX64 T1PA51 A0A224XJN9 A0A026WK75 A0A2R7VP63 A0A3L8DBV3 A0A1L8DRA4 A0A2P8YFD4 A0A085LW55 A0A1L8D8Q4
PDB
2VZN
E-value=2.00883e-19,
Score=232
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.995557
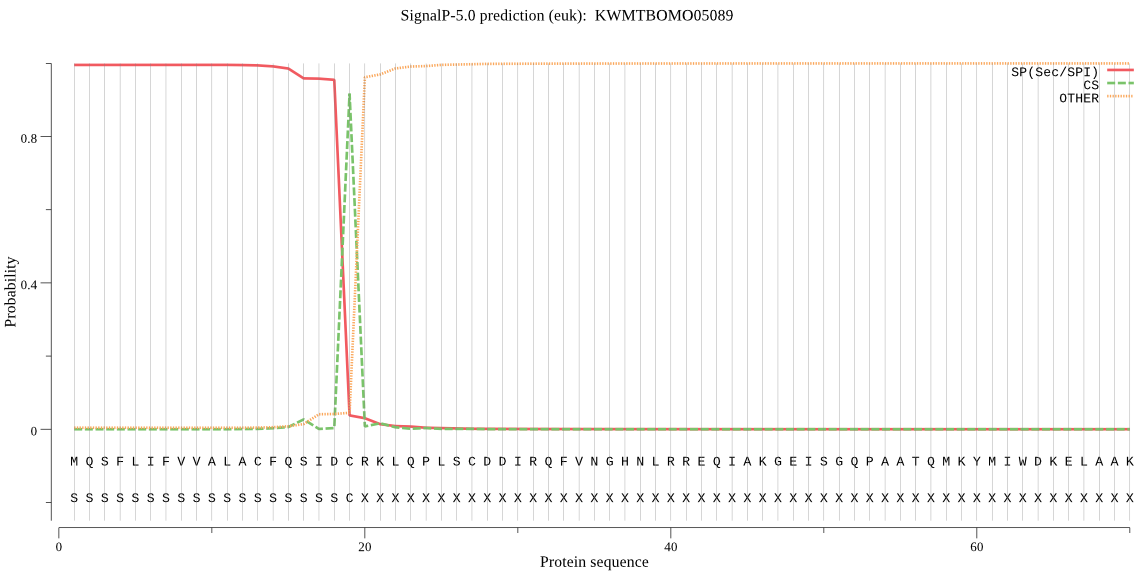
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01986
Exp number, first 60 AAs:
0.0055
Total prob of N-in:
0.01599
outside
1 - 216
Population Genetic Test Statistics
Pi
235.445122
Theta
174.159204
Tajima's D
1.076922
CLR
0
CSRT
0.67911604419779
Interpretation
Uncertain
