Gene
KWMTBOMO05078 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012550
Annotation
signal_peptidase_18_kDa_subunit_[Bombyx_mori]
Full name
Signal peptidase complex catalytic subunit SEC11
Location in the cell
Cytoplasmic Reliability : 1.102 PlasmaMembrane Reliability : 1.396
Sequence
CDS
ATGTTGGAAAGTCTATTTGATGATGTGCGGCGAATGAACAAGCGGCAGTTCATGTACCAAGTGTTAAGCTTTGGTATGATAGTGTCTTCTGCCTTGATGATATGGAAAGGTTTGATGGTAGTAACAGGAAGTGAAAGCCCAATAGTAGTAGTACTGTCAGGCAGTATGGAGCCAGCTTTTCACAGAGGAGACTTGCTGTTTTTAACCAATTACCCTGAGGAGCCTGTCCGCGTTGGAGAAATTGTTGTCTTCAAAGTAGAGGGCCGGGATATCCCGATTGTTCACAGAGTATTGAAACTACATGAAAAGAACAATGGCACAGTAAAGTTCTTAACCAAAGGTGACAATAATAGCGTTGATGACAGAGGCTTGTATGCCCAGGGCCAACTGTGGCTCACTAAAAAAGATGTGGTTGGCCGTGCAAGAGGATTTTTGCCATATGTTGGAATGGTCACCATATATATGAATGAATATCCTAAGTTTAAGTTTGCTGTGCTGGCTTGTCTTGCAATTTATGTATTAGTACACAGGGAGTGA
Protein
MLESLFDDVRRMNKRQFMYQVLSFGMIVSSALMIWKGLMVVTGSESPIVVVLSGSMEPAFHRGDLLFLTNYPEEPVRVGEIVVFKVEGRDIPIVHRVLKLHEKNNGTVKFLTKGDNNSVDDRGLYAQGQLWLTKKDVVGRARGFLPYVGMVTIYMNEYPKFKFAVLACLAIYVLVHRE
Summary
Catalytic Activity
Cleavage of hydrophobic, N-terminal signal or leader sequences from secreted and periplasmic proteins.
Subunit
Component of the signal peptidase complex.
Similarity
Belongs to the peptidase S26B family.
Uniprot
Q2F5L1
A0A1E1WSK5
A0A194RNT8
A0A212EH79
I4DKG1
S4PI45
+ More
A0A2P8ZF68 A0A1Y1NFT5 A0A067RKB9 J3JW13 D6WCL9 N6T7X5 V5IA04 E2A6R7 A0A026WQ02 E9IZM2 E2BXM5 A0A0T6AYL7 F4WDI9 A0A195FRW3 A0A158NIP3 A0A151WI65 A0A151J5T1 R9UE57 A0A1B6DH21 A0A0P4VW34 R4G542 A0A224XLW5 E2J7E8 A0A069DPF7 A0A0A9Y9J6 A0A1W4WW12 A0A0N0BHK2 A0A1B6GV79 A0A1B6KQV8 A0A1J1J634 A0A2H1WNW7 A0A2H8TF19 A0A154P703 C4WT12 A0A2S2PGI3 A0A1L8DUQ9 A0A023FB37 H9JSN9 A0A1D2ND94 A0A2A3E975 V9IJF5 A0A088A934 A0A212EJR7 A0A1B6H5H5 E0VAM4 U5EVP6 A0A0L7RHP6 A0A0K8TQ36 D3PJ83 C1C2A4 A0A023EI51 E9GGE4 A0A0P5RRX4 A0A210QEZ9 C1BS39 A0A182N6K6 A0A2T7PSJ2 C1BVD8 V3ZR90 A0A182TNY9 C3ZJI0 B4NA20 B4KBK0 B4LZ46 A0A182P2Y4 A0A182UUD7 A0A182LC10 A0A182X8B3 A0T4G0 Q06DK2 A0A182HIW8 A0A084VXC7 A0A170YFH8 A0A0L0BUU5 A0A1S3D8K9 A0A182Q9Q2 A0A182XVR1 A0A0P5FIB0 A0A2L2XV54 B4JGV9 A0A1L8EFB7 A7S5L9 A0A1I8P0I9 B3M0S1 A0A182W0B6 A0A087UYK2 A0A1A9XSD1 A0A1A9UNU2 A0A1B0BLX8 D3TNE7 T1ISQ2 A0A1A9WHT6 Q3YMT3 Q3YMT4
A0A2P8ZF68 A0A1Y1NFT5 A0A067RKB9 J3JW13 D6WCL9 N6T7X5 V5IA04 E2A6R7 A0A026WQ02 E9IZM2 E2BXM5 A0A0T6AYL7 F4WDI9 A0A195FRW3 A0A158NIP3 A0A151WI65 A0A151J5T1 R9UE57 A0A1B6DH21 A0A0P4VW34 R4G542 A0A224XLW5 E2J7E8 A0A069DPF7 A0A0A9Y9J6 A0A1W4WW12 A0A0N0BHK2 A0A1B6GV79 A0A1B6KQV8 A0A1J1J634 A0A2H1WNW7 A0A2H8TF19 A0A154P703 C4WT12 A0A2S2PGI3 A0A1L8DUQ9 A0A023FB37 H9JSN9 A0A1D2ND94 A0A2A3E975 V9IJF5 A0A088A934 A0A212EJR7 A0A1B6H5H5 E0VAM4 U5EVP6 A0A0L7RHP6 A0A0K8TQ36 D3PJ83 C1C2A4 A0A023EI51 E9GGE4 A0A0P5RRX4 A0A210QEZ9 C1BS39 A0A182N6K6 A0A2T7PSJ2 C1BVD8 V3ZR90 A0A182TNY9 C3ZJI0 B4NA20 B4KBK0 B4LZ46 A0A182P2Y4 A0A182UUD7 A0A182LC10 A0A182X8B3 A0T4G0 Q06DK2 A0A182HIW8 A0A084VXC7 A0A170YFH8 A0A0L0BUU5 A0A1S3D8K9 A0A182Q9Q2 A0A182XVR1 A0A0P5FIB0 A0A2L2XV54 B4JGV9 A0A1L8EFB7 A7S5L9 A0A1I8P0I9 B3M0S1 A0A182W0B6 A0A087UYK2 A0A1A9XSD1 A0A1A9UNU2 A0A1B0BLX8 D3TNE7 T1ISQ2 A0A1A9WHT6 Q3YMT3 Q3YMT4
EC Number
3.4.21.89
Pubmed
26354079
22118469
22651552
23622113
29403074
28004739
+ More
24845553 22516182 18362917 19820115 23537049 20798317 24508170 30249741 21282665 21719571 21347285 24467622 27129103 26334808 25401762 26823975 25474469 19121390 27289101 20566863 26369729 24945155 26483478 21292972 28812685 23254933 18563158 17994087 20966253 12364791 14747013 17210077 17244545 24438588 26108605 25244985 26561354 17615350 20353571 15917496
24845553 22516182 18362917 19820115 23537049 20798317 24508170 30249741 21282665 21719571 21347285 24467622 27129103 26334808 25401762 26823975 25474469 19121390 27289101 20566863 26369729 24945155 26483478 21292972 28812685 23254933 18563158 17994087 20966253 12364791 14747013 17210077 17244545 24438588 26108605 25244985 26561354 17615350 20353571 15917496
EMBL
DQ311412
ABD36356.1
GDQN01001025
JAT90029.1
KQ459896
KPJ19503.1
+ More
AGBW02014953 OWR40832.1 AK401779 KQ459606 BAM18401.1 KPI91321.1 GAIX01005545 JAA87015.1 PYGN01000075 PSN55146.1 GEZM01003462 JAV96794.1 KK852630 KDR19862.1 BT127431 AEE62393.1 KQ971317 EEZ97812.1 APGK01048489 KB741112 KB632184 ENN73808.1 ERL89640.1 GALX01002030 JAB66436.1 GL437197 EFN70861.1 KK107152 QOIP01000007 EZA57174.1 RLU20654.1 GL767232 EFZ13940.1 GL451281 EFN79543.1 LJIG01022561 KRT79918.1 GL888087 EGI67769.1 KQ981285 KYN43171.1 ADTU01017036 KQ983089 KYQ47527.1 KQ979945 KYN18569.1 KC576515 AGN53399.1 GEDC01012297 JAS25001.1 GDKW01001705 JAI54890.1 ACPB03018001 GAHY01000815 JAA76695.1 GFTR01002961 JAW13465.1 HP429366 ADN29866.1 GBGD01003169 JAC85720.1 GBHO01015841 GBRD01003562 GDHC01005585 JAG27763.1 JAG62259.1 JAQ13044.1 KQ435753 KOX76114.1 GECZ01003440 JAS66329.1 GEBQ01026151 JAT13826.1 CVRI01000073 CRL07867.1 ODYU01009595 SOQ54134.1 GFXV01000869 MBW12674.1 KQ434829 KZC07715.1 ABLF02035666 AK340416 BAH71032.1 GGMR01015920 MBY28539.1 GFDF01003914 JAV10170.1 GBBI01000020 JAC18692.1 BABH01032096 LJIJ01000082 ODN03212.1 KZ288318 PBC28278.1 JR050379 AEY61258.1 AGBW02014409 OWR41710.1 GECU01037747 JAS69959.1 DS235012 EEB10430.1 GANO01001782 JAB58089.1 KQ414590 KOC70318.1 GDAI01001350 JAI16253.1 BT121689 HACA01007091 ADD38619.1 CDW24452.1 BT080983 ACO15407.1 JXUM01101939 GAPW01004831 KQ564686 JAC08767.1 KXJ71898.1 GL732543 EFX81390.1 GDIQ01103913 GDIP01116465 LRGB01000725 JAL47813.1 JAL87249.1 KZS16330.1 NEDP02003952 OWF47322.1 BT077418 ACO11842.1 PZQS01000002 PVD36388.1 BT078567 ACO12991.1 KB203331 ESO85075.1 GG666632 EEN47322.1 CH964232 EDW81775.1 CH933806 EDW13667.1 CH940650 EDW67053.1 AAAB01008879 EAA08439.2 DQ910323 ABI83745.1 APCN01002097 ATLV01017955 KE525202 KFB42621.1 GEMB01003348 JAR99866.1 JRES01001300 KNC23807.1 AXCN02000288 GDIQ01256785 JAJ94939.1 IAAA01008391 IAAA01008392 IAAA01008393 IAAA01008394 IAAA01008395 LAA00026.1 CH916369 EDV93737.1 GFDG01001377 JAV17422.1 DS469583 EDO41033.1 CH902617 EDV42086.1 KK122296 KFM82441.1 JXJN01016598 CCAG010011328 EZ422949 ADD19225.1 JH431446 DQ062793 AAY56666.1 DQ062792 AAY56665.1
AGBW02014953 OWR40832.1 AK401779 KQ459606 BAM18401.1 KPI91321.1 GAIX01005545 JAA87015.1 PYGN01000075 PSN55146.1 GEZM01003462 JAV96794.1 KK852630 KDR19862.1 BT127431 AEE62393.1 KQ971317 EEZ97812.1 APGK01048489 KB741112 KB632184 ENN73808.1 ERL89640.1 GALX01002030 JAB66436.1 GL437197 EFN70861.1 KK107152 QOIP01000007 EZA57174.1 RLU20654.1 GL767232 EFZ13940.1 GL451281 EFN79543.1 LJIG01022561 KRT79918.1 GL888087 EGI67769.1 KQ981285 KYN43171.1 ADTU01017036 KQ983089 KYQ47527.1 KQ979945 KYN18569.1 KC576515 AGN53399.1 GEDC01012297 JAS25001.1 GDKW01001705 JAI54890.1 ACPB03018001 GAHY01000815 JAA76695.1 GFTR01002961 JAW13465.1 HP429366 ADN29866.1 GBGD01003169 JAC85720.1 GBHO01015841 GBRD01003562 GDHC01005585 JAG27763.1 JAG62259.1 JAQ13044.1 KQ435753 KOX76114.1 GECZ01003440 JAS66329.1 GEBQ01026151 JAT13826.1 CVRI01000073 CRL07867.1 ODYU01009595 SOQ54134.1 GFXV01000869 MBW12674.1 KQ434829 KZC07715.1 ABLF02035666 AK340416 BAH71032.1 GGMR01015920 MBY28539.1 GFDF01003914 JAV10170.1 GBBI01000020 JAC18692.1 BABH01032096 LJIJ01000082 ODN03212.1 KZ288318 PBC28278.1 JR050379 AEY61258.1 AGBW02014409 OWR41710.1 GECU01037747 JAS69959.1 DS235012 EEB10430.1 GANO01001782 JAB58089.1 KQ414590 KOC70318.1 GDAI01001350 JAI16253.1 BT121689 HACA01007091 ADD38619.1 CDW24452.1 BT080983 ACO15407.1 JXUM01101939 GAPW01004831 KQ564686 JAC08767.1 KXJ71898.1 GL732543 EFX81390.1 GDIQ01103913 GDIP01116465 LRGB01000725 JAL47813.1 JAL87249.1 KZS16330.1 NEDP02003952 OWF47322.1 BT077418 ACO11842.1 PZQS01000002 PVD36388.1 BT078567 ACO12991.1 KB203331 ESO85075.1 GG666632 EEN47322.1 CH964232 EDW81775.1 CH933806 EDW13667.1 CH940650 EDW67053.1 AAAB01008879 EAA08439.2 DQ910323 ABI83745.1 APCN01002097 ATLV01017955 KE525202 KFB42621.1 GEMB01003348 JAR99866.1 JRES01001300 KNC23807.1 AXCN02000288 GDIQ01256785 JAJ94939.1 IAAA01008391 IAAA01008392 IAAA01008393 IAAA01008394 IAAA01008395 LAA00026.1 CH916369 EDV93737.1 GFDG01001377 JAV17422.1 DS469583 EDO41033.1 CH902617 EDV42086.1 KK122296 KFM82441.1 JXJN01016598 CCAG010011328 EZ422949 ADD19225.1 JH431446 DQ062793 AAY56666.1 DQ062792 AAY56665.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000245037
UP000027135
UP000007266
+ More
UP000019118 UP000030742 UP000000311 UP000053097 UP000279307 UP000008237 UP000007755 UP000078541 UP000005205 UP000075809 UP000078492 UP000015103 UP000192223 UP000053105 UP000183832 UP000076502 UP000007819 UP000005204 UP000094527 UP000242457 UP000005203 UP000009046 UP000053825 UP000069940 UP000249989 UP000000305 UP000076858 UP000242188 UP000075884 UP000245119 UP000030746 UP000075902 UP000001554 UP000007798 UP000009192 UP000008792 UP000075885 UP000075903 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000030765 UP000037069 UP000079169 UP000075886 UP000076408 UP000001070 UP000001593 UP000095300 UP000007801 UP000075920 UP000054359 UP000092443 UP000078200 UP000092460 UP000092444 UP000091820
UP000019118 UP000030742 UP000000311 UP000053097 UP000279307 UP000008237 UP000007755 UP000078541 UP000005205 UP000075809 UP000078492 UP000015103 UP000192223 UP000053105 UP000183832 UP000076502 UP000007819 UP000005204 UP000094527 UP000242457 UP000005203 UP000009046 UP000053825 UP000069940 UP000249989 UP000000305 UP000076858 UP000242188 UP000075884 UP000245119 UP000030746 UP000075902 UP000001554 UP000007798 UP000009192 UP000008792 UP000075885 UP000075903 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000030765 UP000037069 UP000079169 UP000075886 UP000076408 UP000001070 UP000001593 UP000095300 UP000007801 UP000075920 UP000054359 UP000092443 UP000078200 UP000092460 UP000092444 UP000091820
Interpro
IPR019756
Pept_S26A_signal_pept_1_Ser-AS
+ More
IPR037710 SEC11C
IPR015927 Peptidase_S24_S26A/B/C
IPR019758 Pept_S26A_signal_pept_1_CS
IPR036286 LexA/Signal_pep-like_sf
IPR001733 Peptidase_S26B
IPR037712 SEC11A
IPR023318 Ub_act_enz_dom_a_sf
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR030468 Uba3
IPR014929 E2_binding
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR037710 SEC11C
IPR015927 Peptidase_S24_S26A/B/C
IPR019758 Pept_S26A_signal_pept_1_CS
IPR036286 LexA/Signal_pep-like_sf
IPR001733 Peptidase_S26B
IPR037712 SEC11A
IPR023318 Ub_act_enz_dom_a_sf
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR030468 Uba3
IPR014929 E2_binding
IPR033127 UBQ-activ_enz_E1_Cys_AS
Gene 3D
ProteinModelPortal
Q2F5L1
A0A1E1WSK5
A0A194RNT8
A0A212EH79
I4DKG1
S4PI45
+ More
A0A2P8ZF68 A0A1Y1NFT5 A0A067RKB9 J3JW13 D6WCL9 N6T7X5 V5IA04 E2A6R7 A0A026WQ02 E9IZM2 E2BXM5 A0A0T6AYL7 F4WDI9 A0A195FRW3 A0A158NIP3 A0A151WI65 A0A151J5T1 R9UE57 A0A1B6DH21 A0A0P4VW34 R4G542 A0A224XLW5 E2J7E8 A0A069DPF7 A0A0A9Y9J6 A0A1W4WW12 A0A0N0BHK2 A0A1B6GV79 A0A1B6KQV8 A0A1J1J634 A0A2H1WNW7 A0A2H8TF19 A0A154P703 C4WT12 A0A2S2PGI3 A0A1L8DUQ9 A0A023FB37 H9JSN9 A0A1D2ND94 A0A2A3E975 V9IJF5 A0A088A934 A0A212EJR7 A0A1B6H5H5 E0VAM4 U5EVP6 A0A0L7RHP6 A0A0K8TQ36 D3PJ83 C1C2A4 A0A023EI51 E9GGE4 A0A0P5RRX4 A0A210QEZ9 C1BS39 A0A182N6K6 A0A2T7PSJ2 C1BVD8 V3ZR90 A0A182TNY9 C3ZJI0 B4NA20 B4KBK0 B4LZ46 A0A182P2Y4 A0A182UUD7 A0A182LC10 A0A182X8B3 A0T4G0 Q06DK2 A0A182HIW8 A0A084VXC7 A0A170YFH8 A0A0L0BUU5 A0A1S3D8K9 A0A182Q9Q2 A0A182XVR1 A0A0P5FIB0 A0A2L2XV54 B4JGV9 A0A1L8EFB7 A7S5L9 A0A1I8P0I9 B3M0S1 A0A182W0B6 A0A087UYK2 A0A1A9XSD1 A0A1A9UNU2 A0A1B0BLX8 D3TNE7 T1ISQ2 A0A1A9WHT6 Q3YMT3 Q3YMT4
A0A2P8ZF68 A0A1Y1NFT5 A0A067RKB9 J3JW13 D6WCL9 N6T7X5 V5IA04 E2A6R7 A0A026WQ02 E9IZM2 E2BXM5 A0A0T6AYL7 F4WDI9 A0A195FRW3 A0A158NIP3 A0A151WI65 A0A151J5T1 R9UE57 A0A1B6DH21 A0A0P4VW34 R4G542 A0A224XLW5 E2J7E8 A0A069DPF7 A0A0A9Y9J6 A0A1W4WW12 A0A0N0BHK2 A0A1B6GV79 A0A1B6KQV8 A0A1J1J634 A0A2H1WNW7 A0A2H8TF19 A0A154P703 C4WT12 A0A2S2PGI3 A0A1L8DUQ9 A0A023FB37 H9JSN9 A0A1D2ND94 A0A2A3E975 V9IJF5 A0A088A934 A0A212EJR7 A0A1B6H5H5 E0VAM4 U5EVP6 A0A0L7RHP6 A0A0K8TQ36 D3PJ83 C1C2A4 A0A023EI51 E9GGE4 A0A0P5RRX4 A0A210QEZ9 C1BS39 A0A182N6K6 A0A2T7PSJ2 C1BVD8 V3ZR90 A0A182TNY9 C3ZJI0 B4NA20 B4KBK0 B4LZ46 A0A182P2Y4 A0A182UUD7 A0A182LC10 A0A182X8B3 A0T4G0 Q06DK2 A0A182HIW8 A0A084VXC7 A0A170YFH8 A0A0L0BUU5 A0A1S3D8K9 A0A182Q9Q2 A0A182XVR1 A0A0P5FIB0 A0A2L2XV54 B4JGV9 A0A1L8EFB7 A7S5L9 A0A1I8P0I9 B3M0S1 A0A182W0B6 A0A087UYK2 A0A1A9XSD1 A0A1A9UNU2 A0A1B0BLX8 D3TNE7 T1ISQ2 A0A1A9WHT6 Q3YMT3 Q3YMT4
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
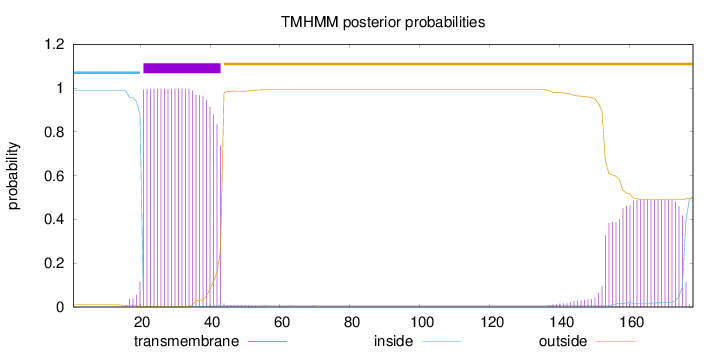
Length:
178
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
33.60669
Exp number, first 60 AAs:
22.48245
Total prob of N-in:
0.99018
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 178
Population Genetic Test Statistics
Pi
263.180191
Theta
185.773245
Tajima's D
1.22337
CLR
0
CSRT
0.722413879306035
Interpretation
Uncertain
