Pre Gene Modal
BGIBMGA012550
Annotation
PREDICTED:_NEDD8-activating_enzyme_E1_catalytic_subunit_[Amyelois_transitella]
Full name
Signal peptidase complex catalytic subunit SEC11
+ More
NEDD8-activating enzyme E1 catalytic subunit
NEDD8-activating enzyme E1 catalytic subunit
Alternative Name
NEDD8-activating enzyme E1C
Ubiquitin-activating enzyme E1C
Ubiquitin-like modifier-activating enzyme 3
Ubiquitin-activating enzyme E1C
Ubiquitin-like modifier-activating enzyme 3
Location in the cell
Cytoplasmic Reliability : 1.656
Sequence
CDS
ATGACTTCAGAATCTCAAACGGCTGATTATCAAAATAGGAGATGGCAACATATAAGGAAGCTGCTGGAAAGATCTGGTCCTTTCTGCCATCCAGATTTTGAAGCTTCAGCAGAAATTCTAAATTTCATCATGAACTGTTGTAAAGTTCTCATTGTTGGGGCAGGTGGTCTTGGTTGTGAATTATTGAAAGATTTAGCCTTAATGGGCTTCAAAAAGTTACATATTGTAGATATGGATACAATAGAACTATCAAATTTAAACCGGCAATTTTTGTTCAGAAGAAATGATATTGGCCTACCTAAAGCTAAATGTGCTGTGGAATTTGTCAATAAAAGAGTTCCTGGGTGTGAAGCTGTAGCTCACCACTGTGCCATTCAGGACCTTGATGAAGATTTTTATAGACAATTTCACATTGTTGTTTGTGGGCTTGACTCAATTATTGCTCGGAGATGGCTTAATGGAATGCTGGTATCCTTGTTGCAGTACAATGATGACAGAAGTTTGGACCAGAGTAGTGTAATTCCAATGGTGGATGGAGGTACTGAGGGATTTAAAGGCAATGCTAGAGTTATATTACCTGGCATGAGTGCATGTATTGAGTGTACTTTAGATCTATATCCCCCACAAAAAACTTTCCCTTTGTGCACCATTGCTAACACTCCAAGATTACCAGAACATTGTATCGAATATGTTAAAGTAATTCAATGGGCAAAGGAGAATCCATGGGGTAGTTCAACAACTTTAGATGGTGATGATCCACAGCATGTCGTGTGGGTGTATGAAAAGGCCCAAGAAAGGGCAATGAAGCATGGGATTGCTAATGTTACATACAGGTTGACCCAAGGAGTTTTAAAGAACATTATACCTGCTGTTGCCAGTACTAATGCTGCTATAGCTGCTGCTTGTGCTACAGAGGTATTTAAACTAGCTTCATCTTGTTGTGTGAATATGAACAACTACATGGTACTGAATATGGCTGATGGTGTGTATACATATACATTTAGTGCAGAGAAAAGGCCTGATTGCGTTGCTTGTTGCAATTCTACTAGGGTTATGAAAATCGATCCTGATGCTACATTACAATATATTTATACAAAACTTTGCGAAGATCAAGCTTATTTGATGAAGAGTCCAGGCATAACCACTGTAATTAATGGATGCAATAAAACATTATACATGTCATCTATTAAGAGCATTGAAGAAAGGACAAAAGATAATTTAAAGAAGAAATTAACAGATTTGGGTTTGTTTAATGGTGCTGATATATTAGTAGCAGATGTGACTACTCCTAACACCATAACCATAAAATTAAAATTCGAAACTCTTGATGTTGATATGACTAAATAA
Protein
MTSESQTADYQNRRWQHIRKLLERSGPFCHPDFEASAEILNFIMNCCKVLIVGAGGLGCELLKDLALMGFKKLHIVDMDTIELSNLNRQFLFRRNDIGLPKAKCAVEFVNKRVPGCEAVAHHCAIQDLDEDFYRQFHIVVCGLDSIIARRWLNGMLVSLLQYNDDRSLDQSSVIPMVDGGTEGFKGNARVILPGMSACIECTLDLYPPQKTFPLCTIANTPRLPEHCIEYVKVIQWAKENPWGSSTTLDGDDPQHVVWVYEKAQERAMKHGIANVTYRLTQGVLKNIIPAVASTNAAIAAACATEVFKLASSCCVNMNNYMVLNMADGVYTYTFSAEKRPDCVACCNSTRVMKIDPDATLQYIYTKLCEDQAYLMKSPGITTVINGCNKTLYMSSIKSIEERTKDNLKKKLTDLGLFNGADILVADVTTPNTITIKLKFETLDVDMTK
Summary
Description
Catalytic subunit of the dimeric uba3-nae1 E1 enzyme. E1 activates nedd8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a nedd8-uba3 thioester and free AMP. E1 finally transfers nedd8 to the catalytic cysteine of ube2m (By similarity).
Catalytic Activity
Cleavage of hydrophobic, N-terminal signal or leader sequences from secreted and periplasmic proteins.
Subunit
Component of the signal peptidase complex.
Heterodimer of uba3 and nae1. Interacts with nedd8, ube2f and ube2m (By similarity).
Heterodimer of uba3 and nae1. Interacts with nedd8, ube2f and ube2m (By similarity).
Miscellaneous
Arg-210 acts as a selectivity gate, preventing misactivation of ubiquitin by this NEDD8-specific E1 complex.
Similarity
Belongs to the peptidase S26B family.
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the ubiquitin-activating E1 family. UBA3 subfamily.
Belongs to the gastrin/cholecystokinin family.
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the ubiquitin-activating E1 family. UBA3 subfamily.
Belongs to the gastrin/cholecystokinin family.
Keywords
ATP-binding
Complete proteome
Ligase
Nucleotide-binding
Reference proteome
Ubl conjugation pathway
Feature
chain NEDD8-activating enzyme E1 catalytic subunit
Uniprot
A0A3S2M6F9
S4P9S9
A0A2H1WM38
A0A194RQ24
A0A194PJQ2
I4DQ99
+ More
H9JSN9 W5JTX7 A0A2M4ANZ9 Q16MG4 A0A182YFM3 A0A182WIJ7 A0A182FCF3 A0A182N898 A0A182VB22 A0A182GNT3 U5EV81 A0A3F2YST0 A0A182RTM1 A0A182U882 A0A182MB21 A0A182JUE6 A0A1B0DMH4 B0XB55 A0A182LBA9 B0X7X9 Q7Q9Q6 A0A182X489 A0A1Q3F3G9 A0A1Y1KEA0 A0A182PZE4 A0A1L8DT78 A0A1B6F1D8 A0A1B6CKV5 A0A1W4X7L3 A0A0A9YQ13 A0A0N7Z926 A0A088ADR7 A0A2A3ENJ2 A0A2J7QDH8 A0A1B6KC19 A0A2R7W4V1 D7EIA6 A0A2P8XTG1 A0A1B6K7X0 A0A1B6GJQ5 E2B846 A0A067RGR8 E0VB43 A0A0K8UAE4 E2ASD8 A0A1I8MBH8 T1PHU3 A0A1I8PPI0 A0A1S3KBU9 A0A1S3KBB4 A0A034WB58 A0A0A1XEV5 A0A0M8ZP40 B2GSP4 Q7ZVX6 A0A0J7KST6 C3YVJ7 W8BKT5 F4WSY0 A0A210R4K4 A0A0B7B6L3 A0A1A8FKP0 F6YX67 A0A1W2WGT2 A0A151WTU5 A0A3B5B5I1 A0A151J534 A0A3B3QW48 H3D4U0 A0A3B3QVH4 A0A1A8KAY1 A0A3Q3NH78 A0A3Q1K8U5 Q4S5R8 A0A3B1K7Q7 A0A1J1IIR9 A0A3B3QVK3 A0A3B4VC00 A0A1A8R0Z4 A0A1A8LW05 A0A1A8BVI5 A0A3B1J473 A0A1A7YN26 A0A1A8VJA2 A0A3S0ZWF3 A0A3Q3T4A5 A0A1W5APB5 A0A1W5AX36 A0A0K8RSG2 A0A026WAQ0 A0A1W5APF5 A0A3B3BEG2 A0A3Q3F630
H9JSN9 W5JTX7 A0A2M4ANZ9 Q16MG4 A0A182YFM3 A0A182WIJ7 A0A182FCF3 A0A182N898 A0A182VB22 A0A182GNT3 U5EV81 A0A3F2YST0 A0A182RTM1 A0A182U882 A0A182MB21 A0A182JUE6 A0A1B0DMH4 B0XB55 A0A182LBA9 B0X7X9 Q7Q9Q6 A0A182X489 A0A1Q3F3G9 A0A1Y1KEA0 A0A182PZE4 A0A1L8DT78 A0A1B6F1D8 A0A1B6CKV5 A0A1W4X7L3 A0A0A9YQ13 A0A0N7Z926 A0A088ADR7 A0A2A3ENJ2 A0A2J7QDH8 A0A1B6KC19 A0A2R7W4V1 D7EIA6 A0A2P8XTG1 A0A1B6K7X0 A0A1B6GJQ5 E2B846 A0A067RGR8 E0VB43 A0A0K8UAE4 E2ASD8 A0A1I8MBH8 T1PHU3 A0A1I8PPI0 A0A1S3KBU9 A0A1S3KBB4 A0A034WB58 A0A0A1XEV5 A0A0M8ZP40 B2GSP4 Q7ZVX6 A0A0J7KST6 C3YVJ7 W8BKT5 F4WSY0 A0A210R4K4 A0A0B7B6L3 A0A1A8FKP0 F6YX67 A0A1W2WGT2 A0A151WTU5 A0A3B5B5I1 A0A151J534 A0A3B3QW48 H3D4U0 A0A3B3QVH4 A0A1A8KAY1 A0A3Q3NH78 A0A3Q1K8U5 Q4S5R8 A0A3B1K7Q7 A0A1J1IIR9 A0A3B3QVK3 A0A3B4VC00 A0A1A8R0Z4 A0A1A8LW05 A0A1A8BVI5 A0A3B1J473 A0A1A7YN26 A0A1A8VJA2 A0A3S0ZWF3 A0A3Q3T4A5 A0A1W5APB5 A0A1W5AX36 A0A0K8RSG2 A0A026WAQ0 A0A1W5APF5 A0A3B3BEG2 A0A3Q3F630
EC Number
3.4.21.89
6.2.1.-
6.2.1.-
Pubmed
23622113
26354079
22651552
19121390
20920257
23761445
+ More
17510324 25244985 26483478 20966253 12364791 14747013 17210077 28004739 25401762 26823975 27129103 18362917 19820115 29403074 20798317 24845553 20566863 25315136 25348373 25830018 18563158 24495485 21719571 28812685 12481130 15114417 29240929 15496914 25329095 25727380 24508170 29451363
17510324 25244985 26483478 20966253 12364791 14747013 17210077 28004739 25401762 26823975 27129103 18362917 19820115 29403074 20798317 24845553 20566863 25315136 25348373 25830018 18563158 24495485 21719571 28812685 12481130 15114417 29240929 15496914 25329095 25727380 24508170 29451363
EMBL
RSAL01000026
RVE52056.1
GAIX01003604
JAA88956.1
ODYU01009595
SOQ54135.1
+ More
KQ459896 KPJ19504.1 KQ459606 KPI91320.1 AK404006 BAM20089.1 BABH01032096 ADMH02000391 ETN66743.1 GGFK01009193 MBW42514.1 CH477862 EAT35524.1 JXUM01077073 KQ562985 KXJ74704.1 GANO01001997 JAB57874.1 APCN01002931 AXCM01004441 AJVK01001342 DS232614 EDS44108.1 DS232468 EDS42198.1 AAAB01008900 EAA09425.1 GFDL01012968 JAV22077.1 GEZM01087693 JAV58530.1 AXCN02000962 GFDF01004428 JAV09656.1 GECZ01025764 JAS44005.1 GEDC01023209 JAS14089.1 GBHO01008502 GBHO01008501 GBRD01002464 GDHC01006004 JAG35102.1 JAG35103.1 JAG63357.1 JAQ12625.1 GDKW01001723 JAI54872.1 KZ288206 PBC33074.1 NEVH01015820 PNF26642.1 GEBQ01030978 JAT08999.1 KK854218 PTY13245.1 DS497675 EFA11744.1 PYGN01001375 PSN35307.1 GECU01000148 JAT07559.1 GECZ01007087 JAS62682.1 GL446282 EFN88138.1 KK852476 KDR23046.1 DS235021 EEB10599.1 GDHF01028746 JAI23568.1 GL442298 EFN63604.1 KA648254 AFP62883.1 GAKP01007587 JAC51365.1 GBXI01004373 JAD09919.1 KQ435944 KOX68215.1 BC165593 AAI65593.1 BC045372 LBMM01003542 KMQ93403.1 GG666557 EEN55720.1 GAMC01008987 JAB97568.1 GL888331 EGI62684.1 NEDP02000459 OWF55844.1 HACG01042084 CEK88949.1 HAEB01012866 HAEC01006753 SBQ59393.1 EAAA01000397 KQ982750 KYQ51264.1 KQ980055 KYN17979.1 HAED01008973 HAEE01009468 SBR29518.1 CAAE01014729 CAG04014.1 CVRI01000054 CRL00088.1 HAEI01005164 HAEH01014100 SBR99123.1 HAEF01009259 HAEG01015664 SBR48154.1 HADZ01006877 HAEA01010240 SBP70818.1 HADW01001255 HADX01009312 SBP31544.1 HADY01015704 HAEJ01019484 SBS59941.1 RQTK01000085 RUS88358.1 GBKC01002339 JAG43731.1 KK107295 EZA53115.1
KQ459896 KPJ19504.1 KQ459606 KPI91320.1 AK404006 BAM20089.1 BABH01032096 ADMH02000391 ETN66743.1 GGFK01009193 MBW42514.1 CH477862 EAT35524.1 JXUM01077073 KQ562985 KXJ74704.1 GANO01001997 JAB57874.1 APCN01002931 AXCM01004441 AJVK01001342 DS232614 EDS44108.1 DS232468 EDS42198.1 AAAB01008900 EAA09425.1 GFDL01012968 JAV22077.1 GEZM01087693 JAV58530.1 AXCN02000962 GFDF01004428 JAV09656.1 GECZ01025764 JAS44005.1 GEDC01023209 JAS14089.1 GBHO01008502 GBHO01008501 GBRD01002464 GDHC01006004 JAG35102.1 JAG35103.1 JAG63357.1 JAQ12625.1 GDKW01001723 JAI54872.1 KZ288206 PBC33074.1 NEVH01015820 PNF26642.1 GEBQ01030978 JAT08999.1 KK854218 PTY13245.1 DS497675 EFA11744.1 PYGN01001375 PSN35307.1 GECU01000148 JAT07559.1 GECZ01007087 JAS62682.1 GL446282 EFN88138.1 KK852476 KDR23046.1 DS235021 EEB10599.1 GDHF01028746 JAI23568.1 GL442298 EFN63604.1 KA648254 AFP62883.1 GAKP01007587 JAC51365.1 GBXI01004373 JAD09919.1 KQ435944 KOX68215.1 BC165593 AAI65593.1 BC045372 LBMM01003542 KMQ93403.1 GG666557 EEN55720.1 GAMC01008987 JAB97568.1 GL888331 EGI62684.1 NEDP02000459 OWF55844.1 HACG01042084 CEK88949.1 HAEB01012866 HAEC01006753 SBQ59393.1 EAAA01000397 KQ982750 KYQ51264.1 KQ980055 KYN17979.1 HAED01008973 HAEE01009468 SBR29518.1 CAAE01014729 CAG04014.1 CVRI01000054 CRL00088.1 HAEI01005164 HAEH01014100 SBR99123.1 HAEF01009259 HAEG01015664 SBR48154.1 HADZ01006877 HAEA01010240 SBP70818.1 HADW01001255 HADX01009312 SBP31544.1 HADY01015704 HAEJ01019484 SBS59941.1 RQTK01000085 RUS88358.1 GBKC01002339 JAG43731.1 KK107295 EZA53115.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000005204
UP000000673
UP000008820
+ More
UP000076408 UP000075920 UP000069272 UP000075884 UP000075903 UP000069940 UP000249989 UP000075840 UP000075900 UP000075902 UP000075883 UP000075881 UP000092462 UP000002320 UP000075882 UP000007062 UP000076407 UP000075886 UP000192223 UP000005203 UP000242457 UP000235965 UP000007266 UP000245037 UP000008237 UP000027135 UP000009046 UP000000311 UP000095301 UP000095300 UP000085678 UP000053105 UP000000437 UP000036403 UP000001554 UP000007755 UP000242188 UP000008144 UP000075809 UP000261400 UP000078492 UP000261540 UP000007303 UP000261640 UP000265040 UP000018467 UP000183832 UP000261420 UP000271974 UP000192224 UP000053097 UP000261560 UP000261660
UP000076408 UP000075920 UP000069272 UP000075884 UP000075903 UP000069940 UP000249989 UP000075840 UP000075900 UP000075902 UP000075883 UP000075881 UP000092462 UP000002320 UP000075882 UP000007062 UP000076407 UP000075886 UP000192223 UP000005203 UP000242457 UP000235965 UP000007266 UP000245037 UP000008237 UP000027135 UP000009046 UP000000311 UP000095301 UP000095300 UP000085678 UP000053105 UP000000437 UP000036403 UP000001554 UP000007755 UP000242188 UP000008144 UP000075809 UP000261400 UP000078492 UP000261540 UP000007303 UP000261640 UP000265040 UP000018467 UP000183832 UP000261420 UP000271974 UP000192224 UP000053097 UP000261560 UP000261660
PRIDE
Interpro
IPR033127
UBQ-activ_enz_E1_Cys_AS
+ More
IPR014929 E2_binding
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR030468 Uba3
IPR023318 Ub_act_enz_dom_a_sf
IPR019758 Pept_S26A_signal_pept_1_CS
IPR036286 LexA/Signal_pep-like_sf
IPR001733 Peptidase_S26B
IPR015927 Peptidase_S24_S26A/B/C
IPR019756 Pept_S26A_signal_pept_1_Ser-AS
IPR029058 AB_hydrolase
IPR013818 Lipase/vitellogenin
IPR033906 Lipase_N
IPR000734 TAG_lipase
IPR000994 Pept_M24
IPR036005 Creatinase/aminopeptidase-like
IPR004545 PA2G4
IPR036388 WH-like_DNA-bd_sf
IPR036390 WH_DNA-bd_sf
IPR013152 Gastrin/cholecystokinin_CS
IPR014929 E2_binding
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR030468 Uba3
IPR023318 Ub_act_enz_dom_a_sf
IPR019758 Pept_S26A_signal_pept_1_CS
IPR036286 LexA/Signal_pep-like_sf
IPR001733 Peptidase_S26B
IPR015927 Peptidase_S24_S26A/B/C
IPR019756 Pept_S26A_signal_pept_1_Ser-AS
IPR029058 AB_hydrolase
IPR013818 Lipase/vitellogenin
IPR033906 Lipase_N
IPR000734 TAG_lipase
IPR000994 Pept_M24
IPR036005 Creatinase/aminopeptidase-like
IPR004545 PA2G4
IPR036388 WH-like_DNA-bd_sf
IPR036390 WH_DNA-bd_sf
IPR013152 Gastrin/cholecystokinin_CS
SUPFAM
Gene 3D
ProteinModelPortal
A0A3S2M6F9
S4P9S9
A0A2H1WM38
A0A194RQ24
A0A194PJQ2
I4DQ99
+ More
H9JSN9 W5JTX7 A0A2M4ANZ9 Q16MG4 A0A182YFM3 A0A182WIJ7 A0A182FCF3 A0A182N898 A0A182VB22 A0A182GNT3 U5EV81 A0A3F2YST0 A0A182RTM1 A0A182U882 A0A182MB21 A0A182JUE6 A0A1B0DMH4 B0XB55 A0A182LBA9 B0X7X9 Q7Q9Q6 A0A182X489 A0A1Q3F3G9 A0A1Y1KEA0 A0A182PZE4 A0A1L8DT78 A0A1B6F1D8 A0A1B6CKV5 A0A1W4X7L3 A0A0A9YQ13 A0A0N7Z926 A0A088ADR7 A0A2A3ENJ2 A0A2J7QDH8 A0A1B6KC19 A0A2R7W4V1 D7EIA6 A0A2P8XTG1 A0A1B6K7X0 A0A1B6GJQ5 E2B846 A0A067RGR8 E0VB43 A0A0K8UAE4 E2ASD8 A0A1I8MBH8 T1PHU3 A0A1I8PPI0 A0A1S3KBU9 A0A1S3KBB4 A0A034WB58 A0A0A1XEV5 A0A0M8ZP40 B2GSP4 Q7ZVX6 A0A0J7KST6 C3YVJ7 W8BKT5 F4WSY0 A0A210R4K4 A0A0B7B6L3 A0A1A8FKP0 F6YX67 A0A1W2WGT2 A0A151WTU5 A0A3B5B5I1 A0A151J534 A0A3B3QW48 H3D4U0 A0A3B3QVH4 A0A1A8KAY1 A0A3Q3NH78 A0A3Q1K8U5 Q4S5R8 A0A3B1K7Q7 A0A1J1IIR9 A0A3B3QVK3 A0A3B4VC00 A0A1A8R0Z4 A0A1A8LW05 A0A1A8BVI5 A0A3B1J473 A0A1A7YN26 A0A1A8VJA2 A0A3S0ZWF3 A0A3Q3T4A5 A0A1W5APB5 A0A1W5AX36 A0A0K8RSG2 A0A026WAQ0 A0A1W5APF5 A0A3B3BEG2 A0A3Q3F630
H9JSN9 W5JTX7 A0A2M4ANZ9 Q16MG4 A0A182YFM3 A0A182WIJ7 A0A182FCF3 A0A182N898 A0A182VB22 A0A182GNT3 U5EV81 A0A3F2YST0 A0A182RTM1 A0A182U882 A0A182MB21 A0A182JUE6 A0A1B0DMH4 B0XB55 A0A182LBA9 B0X7X9 Q7Q9Q6 A0A182X489 A0A1Q3F3G9 A0A1Y1KEA0 A0A182PZE4 A0A1L8DT78 A0A1B6F1D8 A0A1B6CKV5 A0A1W4X7L3 A0A0A9YQ13 A0A0N7Z926 A0A088ADR7 A0A2A3ENJ2 A0A2J7QDH8 A0A1B6KC19 A0A2R7W4V1 D7EIA6 A0A2P8XTG1 A0A1B6K7X0 A0A1B6GJQ5 E2B846 A0A067RGR8 E0VB43 A0A0K8UAE4 E2ASD8 A0A1I8MBH8 T1PHU3 A0A1I8PPI0 A0A1S3KBU9 A0A1S3KBB4 A0A034WB58 A0A0A1XEV5 A0A0M8ZP40 B2GSP4 Q7ZVX6 A0A0J7KST6 C3YVJ7 W8BKT5 F4WSY0 A0A210R4K4 A0A0B7B6L3 A0A1A8FKP0 F6YX67 A0A1W2WGT2 A0A151WTU5 A0A3B5B5I1 A0A151J534 A0A3B3QW48 H3D4U0 A0A3B3QVH4 A0A1A8KAY1 A0A3Q3NH78 A0A3Q1K8U5 Q4S5R8 A0A3B1K7Q7 A0A1J1IIR9 A0A3B3QVK3 A0A3B4VC00 A0A1A8R0Z4 A0A1A8LW05 A0A1A8BVI5 A0A3B1J473 A0A1A7YN26 A0A1A8VJA2 A0A3S0ZWF3 A0A3Q3T4A5 A0A1W5APB5 A0A1W5AX36 A0A0K8RSG2 A0A026WAQ0 A0A1W5APF5 A0A3B3BEG2 A0A3Q3F630
PDB
2NVU
E-value=8.32251e-158,
Score=1429
Ontologies
GO
PANTHER
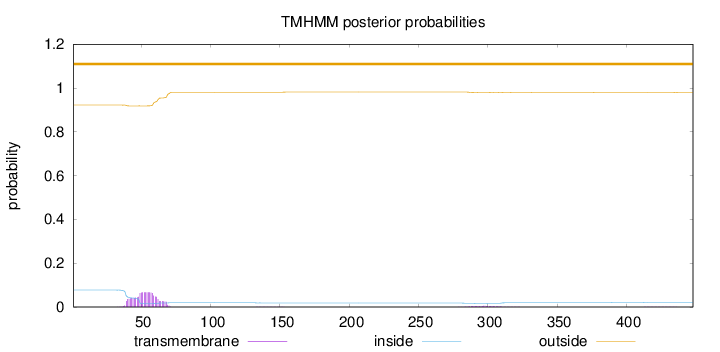
Topology
Subcellular location
Endoplasmic reticulum membrane
Secreted
Secreted
Length:
448
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.5602
Exp number, first 60 AAs:
1.17091
Total prob of N-in:
0.07733
outside
1 - 448
Population Genetic Test Statistics
Pi
4.699553
Theta
12.570161
Tajima's D
-1.704213
CLR
247.156569
CSRT
0.0351982400879956
Interpretation
Uncertain
