Gene
KWMTBOMO05075 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012549
Annotation
H+_transporting_ATP_synthase_beta_subunit_isoform_1_[Bombyx_mori]
Full name
ATP synthase subunit beta
+ More
ATP synthase subunit beta, mitochondrial
ATP synthase subunit beta, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.356 Mitochondrial Reliability : 2.027
Sequence
CDS
ATGTTGGGGGCAATAAGTCGAGTTGGAAGCGGCATTTTAGCCGTTAAATCAGTTGCTGAGAAATCCCTTTCAGAATGTGGAAAAATCGTTGCAGTCAATGCCGTTAACAAGAGGGACTATGCTGCTAAATCTGCAGGAAAAGGCCAAGGTAAGGTTGTTGCCGTTATCGGTGCTGTGGTAGATGTGCAATTCGAAGATAACCTGCCTCCGATTCTAAATGCCCTTGAGGTGCAAAATCGATCTCCCCGCCTCGTACTTGAGGTAGCCCAACATTTGGGTGAGAACACAGTTCGGACCATTGCCATGGACGGTACTGAAGGCTTAGTCCGTGGGCAACCCGTACTAGACTCTGGCTCACCAATTCGTATCCCGGTGGGAGCTGAAACCCTCGGTCGCATCATCAATGTAATCGGCGAACCGATTGACGAGCGTGGTCCCATCCCCACCGACAAGACTGCTGCTATTCATGCTGAAGCTCCAGAGTTTGTCGACATGTCTGTGCAGCAGGAGATTCTCGTAACTGGTATAAAAGTCGTCGATCTGCTCGCTCCTTATGCCAAAGGAGGAAAGATTGGGTTGTTTGGCGGAGCTGGTGTGGGCAAAACTGTATTGATTATGGAACTGATCAACAATGTTGCCAAAGCCCATGGTGGTTACTCTGTGTTTGCTGGAGTAGGAGAGCGTACTCGTGAAGGAAATGATTTATACCACGAGATGATTGAATCTGGTGTGATTTCTCTAAAAGACAAAACATCCAAGGTAGCTCTAGTATATGGTCAAATGAACGAACCTCCTGGTGCCCGTGCTCGTGTTGCCCTTACTGGTCTCACTGTTGCTGAATATTTCCGTGATCAAGAAGGACAGGATGTACTGCTCTTCATTGACAACATTTTCCGTTTCACTCAGGCTGGATCAGAAGTGTCTGCTCTGCTTGGTCGTATCCCATCTGCTGTAGGATATCAGCCTACTCTGGCTACTGACATGGGTACTATGCAGGAAAGAATTACCACCACCAAGAAAGGTTCCATCACATCTGTACAGGCTATTTATGTACCAGCTGATGACTTGACAGATCCTGCTCCAGCTACCACTTTTGCTCACTTGGATGCTACCACTGTACTCTCCAGAGCCATTGCTGAATTGGGTATCTACCCAGCTGTGGATCCTCTTGACTCAACTTCCCGTATCATGGACCCCAATATTATTGGAGCTGAGCACTACAATGTTGCACGTGGAGTTCAGAAAATTCTTCAGGACTACAAATCCCTGCAGGACATTATTGCTATTTTGGGTATGGACGAGTTGTCTGAAGAAGACAAGTTGACAGTGGCACGTGCACGTAAAATTCAGAGGTTCCTCTCACAACCTTTCCAAGTAGCTGAGGTGTTCACTGGACATGCGGGTAAACTAGTACCACTTGAGGAAACTATCAAAGGATTCTCCAAAATTCTAGCAGGGGACTATGATCATTTGCCAGAAGTAGCATTCTACATGGTTGGACCTATTGAGGAGGTTGTGGCCAAAGCTGATACTCTTGCTAAGAATGCTTAA
Protein
MLGAISRVGSGILAVKSVAEKSLSECGKIVAVNAVNKRDYAAKSAGKGQGKVVAVIGAVVDVQFEDNLPPILNALEVQNRSPRLVLEVAQHLGENTVRTIAMDGTEGLVRGQPVLDSGSPIRIPVGAETLGRIINVIGEPIDERGPIPTDKTAAIHAEAPEFVDMSVQQEILVTGIKVVDLLAPYAKGGKIGLFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIESGVISLKDKTSKVALVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAVGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRAIAELGIYPAVDPLDSTSRIMDPNIIGAEHYNVARGVQKILQDYKSLQDIIAILGMDELSEEDKLTVARARKIQRFLSQPFQVAEVFTGHAGKLVPLEETIKGFSKILAGDYDHLPEVAFYMVGPIEEVVAKADTLAKNA
Summary
Description
Produces ATP from ADP in the presence of a proton gradient across the membrane.
Catalytic Activity
ATP + 4 H(+)(in) + H2O = ADP + 5 H(+)(out) + phosphate
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase alpha/beta chains family.
Keywords
ATP synthesis
ATP-binding
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Nucleotide-binding
Reference proteome
Transit peptide
Translocase
Transport
Feature
chain ATP synthase subunit beta, mitochondrial
Uniprot
Q1HPT1
A0A194RTR3
A0A194PE66
I4DM90
K9RZR5
D9ILX6
+ More
Q1HPT0 A0A194RQB3 A0A212EJP1 H9JSP4 I4DMG2 I4DIU5 V9IJZ2 A0A088AMB8 A0A0J7KRQ1 A0A1W4X6X1 A0A154P1L6 D2A4R3 A0A1W6EVQ8 A0A2A3E923 A0A2J7QV00 A0A191UR82 R4WNI7 A0A0N0BJR7 E2C3U0 E2ABW9 A0A2R7VST6 A0A0C9Q3Q8 A0A195BY79 A0A1I9WLI2 A0A067QNJ1 A0A026X0M9 A0A1B6ENI6 V5I8R7 A0A2D1QUC6 A0A0L7KWW5 G9C5G1 A0A195F1S5 A0A0L0BSV2 B4L7D0 A0A158NL77 X1WI47 B4JZN9 A0A232EXM3 A0A023F989 A0A161MK87 Q1ZZQ8 B3N1J9 A0A1L8EHP6 E0W1G3 A0A0M4F807 R4FQF4 L0MQ04 A0A1L8EEU5 D0Z794 X2JH42 B3P9Q0 W0AMY4 Q05825 A0A151JSF9 B4PW41 B4NHL2 J3JX90 A0A3L8DTN4 A0A1I8MLA9 W8BZZ5 T1PAN5 A0A034VA91 A0A1L8EF05 A0A1W4V506 A0A224XET0 B4MEZ1 A0A1P9FG40 U4U6D1 A0A034V7I6 A0A1B6D1T9 A0A1I8NT76 A0A151WHD6 A0A3B0KSV3 Q29CL2 B4H9P4 A0A0A1WKN6 W8BCZ3 A0A0L7RCQ4 U5EUX4 A0A1D1YVH1 A0A1Y1KCU8 A0A195BH40 A0A1L8E9X6 A0A1B0CLT6 A0A1A9V556 D3TRW4 A0A0V0G2S7 A0A1A9ZWH1 A0A1A9YCL3 A0A1B0AQN2 A0A1L8E0V2 A0A1L8E0Y3 A0A1Q3FH39 A0A0A9Y314
Q1HPT0 A0A194RQB3 A0A212EJP1 H9JSP4 I4DMG2 I4DIU5 V9IJZ2 A0A088AMB8 A0A0J7KRQ1 A0A1W4X6X1 A0A154P1L6 D2A4R3 A0A1W6EVQ8 A0A2A3E923 A0A2J7QV00 A0A191UR82 R4WNI7 A0A0N0BJR7 E2C3U0 E2ABW9 A0A2R7VST6 A0A0C9Q3Q8 A0A195BY79 A0A1I9WLI2 A0A067QNJ1 A0A026X0M9 A0A1B6ENI6 V5I8R7 A0A2D1QUC6 A0A0L7KWW5 G9C5G1 A0A195F1S5 A0A0L0BSV2 B4L7D0 A0A158NL77 X1WI47 B4JZN9 A0A232EXM3 A0A023F989 A0A161MK87 Q1ZZQ8 B3N1J9 A0A1L8EHP6 E0W1G3 A0A0M4F807 R4FQF4 L0MQ04 A0A1L8EEU5 D0Z794 X2JH42 B3P9Q0 W0AMY4 Q05825 A0A151JSF9 B4PW41 B4NHL2 J3JX90 A0A3L8DTN4 A0A1I8MLA9 W8BZZ5 T1PAN5 A0A034VA91 A0A1L8EF05 A0A1W4V506 A0A224XET0 B4MEZ1 A0A1P9FG40 U4U6D1 A0A034V7I6 A0A1B6D1T9 A0A1I8NT76 A0A151WHD6 A0A3B0KSV3 Q29CL2 B4H9P4 A0A0A1WKN6 W8BCZ3 A0A0L7RCQ4 U5EUX4 A0A1D1YVH1 A0A1Y1KCU8 A0A195BH40 A0A1L8E9X6 A0A1B0CLT6 A0A1A9V556 D3TRW4 A0A0V0G2S7 A0A1A9ZWH1 A0A1A9YCL3 A0A1B0AQN2 A0A1L8E0V2 A0A1L8E0Y3 A0A1Q3FH39 A0A0A9Y314
EC Number
7.1.2.2
Pubmed
19121390
26354079
22651552
22848670
22118469
18362917
+ More
19820115 27142481 23691247 20798317 27538518 24845553 24508170 26227816 21835214 26108605 17994087 21347285 28648823 25474469 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26334808 8373413 10071211 12537569 17550304 22516182 23537049 30249741 25315136 24495485 25348373 15632085 25830018 28004739 20353571 25401762
19820115 27142481 23691247 20798317 27538518 24845553 24508170 26227816 21835214 26108605 17994087 21347285 28648823 25474469 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26334808 8373413 10071211 12537569 17550304 22516182 23537049 30249741 25315136 24495485 25348373 15632085 25830018 28004739 20353571 25401762
EMBL
BABH01032096
DQ443321
ABF51410.1
KQ459896
KPJ19506.1
KQ459606
+ More
KPI91318.1 AK402408 BAM19030.1 KC018474 AFY62978.1 HM156739 ADJ95799.1 DQ443322 ABF51411.1 KPJ19510.1 AGBW02014409 OWR41713.1 OWR41718.1 BABH01032099 AK402480 BAM19102.1 AK401213 BAM17835.1 KPI91315.1 JR048957 AEY60811.1 LBMM01003998 KMQ92914.1 KQ434796 KZC05747.1 KQ971344 EFA05714.1 KY563404 ARK19813.1 KZ288340 PBC27772.1 NEVH01010480 PNF32408.1 KU365964 ANJ04680.1 AK417201 BAN20416.1 KQ435711 KOX79538.1 GL452364 EFN77326.1 GL438386 EFN69064.1 KK854065 PTY10574.1 GBYB01008703 JAG78470.1 KQ978501 KYM93547.1 KU932366 APA34002.1 KK853123 KDR11062.1 KK107054 EZA61603.1 GECZ01030282 JAS39487.1 GALX01004215 JAB64251.1 KY285053 ATP16156.1 JTDY01004796 KOB67743.1 HQ851402 AEV89780.1 KQ981866 KYN34049.1 JRES01001407 KNC23160.1 CH933813 EDW10924.1 ADTU01019278 ABLF02014736 CH916380 EDV90905.1 NNAY01001745 OXU23047.1 GBBI01000645 JAC18067.1 GEMB01000491 JAS02636.1 DQ416018 ABD76365.1 CH902659 EDV33934.1 GFDG01000562 JAV18237.1 DS235870 EEB19469.1 CP012527 ALC48082.1 ACPB03010146 GAHY01000424 JAA77086.1 AE014135 AGB96571.1 GFDG01001625 JAV17174.1 CM000831 ACY70473.1 AHN58220.1 CH954184 EDV45213.1 KF724682 GBGD01001152 AHE57675.1 JAC87737.1 X71013 AY118367 KQ978530 KYN30286.1 CM000161 EDW99346.1 CH964272 EDW83581.1 APGK01044225 BT127859 KB741021 AEE62821.1 ENN75086.1 QOIP01000004 RLU23662.1 GAMC01011671 JAB94884.1 KA645220 AFP59849.1 GAKP01020469 JAC38483.1 GFDG01001528 JAV17271.1 GFTR01006912 JAW09514.1 CH940665 EDW71092.1 KX357709 AQE30075.1 KB632003 ERL87863.1 GAKP01020468 JAC38484.1 GEDC01017682 JAS19616.1 KQ983120 KYQ47240.1 OUUW01000017 SPP88946.1 CH475455 EAL29273.1 CH479230 EDW36528.1 GBXI01014683 JAC99608.1 GAMC01011672 JAB94883.1 KQ414615 KOC68579.1 GANO01001262 JAB58609.1 GDJX01009324 JAT58612.1 GEZM01088048 JAV58328.1 KQ976488 KYM83491.1 GFDG01003404 JAV15395.1 AJWK01017609 CCAG010007514 EZ424166 ADD20442.1 GECL01003747 JAP02377.1 JXJN01001971 GFDF01001726 JAV12358.1 GFDF01001727 JAV12357.1 GFDL01008177 JAV26868.1 GBHO01017578 JAG26026.1
KPI91318.1 AK402408 BAM19030.1 KC018474 AFY62978.1 HM156739 ADJ95799.1 DQ443322 ABF51411.1 KPJ19510.1 AGBW02014409 OWR41713.1 OWR41718.1 BABH01032099 AK402480 BAM19102.1 AK401213 BAM17835.1 KPI91315.1 JR048957 AEY60811.1 LBMM01003998 KMQ92914.1 KQ434796 KZC05747.1 KQ971344 EFA05714.1 KY563404 ARK19813.1 KZ288340 PBC27772.1 NEVH01010480 PNF32408.1 KU365964 ANJ04680.1 AK417201 BAN20416.1 KQ435711 KOX79538.1 GL452364 EFN77326.1 GL438386 EFN69064.1 KK854065 PTY10574.1 GBYB01008703 JAG78470.1 KQ978501 KYM93547.1 KU932366 APA34002.1 KK853123 KDR11062.1 KK107054 EZA61603.1 GECZ01030282 JAS39487.1 GALX01004215 JAB64251.1 KY285053 ATP16156.1 JTDY01004796 KOB67743.1 HQ851402 AEV89780.1 KQ981866 KYN34049.1 JRES01001407 KNC23160.1 CH933813 EDW10924.1 ADTU01019278 ABLF02014736 CH916380 EDV90905.1 NNAY01001745 OXU23047.1 GBBI01000645 JAC18067.1 GEMB01000491 JAS02636.1 DQ416018 ABD76365.1 CH902659 EDV33934.1 GFDG01000562 JAV18237.1 DS235870 EEB19469.1 CP012527 ALC48082.1 ACPB03010146 GAHY01000424 JAA77086.1 AE014135 AGB96571.1 GFDG01001625 JAV17174.1 CM000831 ACY70473.1 AHN58220.1 CH954184 EDV45213.1 KF724682 GBGD01001152 AHE57675.1 JAC87737.1 X71013 AY118367 KQ978530 KYN30286.1 CM000161 EDW99346.1 CH964272 EDW83581.1 APGK01044225 BT127859 KB741021 AEE62821.1 ENN75086.1 QOIP01000004 RLU23662.1 GAMC01011671 JAB94884.1 KA645220 AFP59849.1 GAKP01020469 JAC38483.1 GFDG01001528 JAV17271.1 GFTR01006912 JAW09514.1 CH940665 EDW71092.1 KX357709 AQE30075.1 KB632003 ERL87863.1 GAKP01020468 JAC38484.1 GEDC01017682 JAS19616.1 KQ983120 KYQ47240.1 OUUW01000017 SPP88946.1 CH475455 EAL29273.1 CH479230 EDW36528.1 GBXI01014683 JAC99608.1 GAMC01011672 JAB94883.1 KQ414615 KOC68579.1 GANO01001262 JAB58609.1 GDJX01009324 JAT58612.1 GEZM01088048 JAV58328.1 KQ976488 KYM83491.1 GFDG01003404 JAV15395.1 AJWK01017609 CCAG010007514 EZ424166 ADD20442.1 GECL01003747 JAP02377.1 JXJN01001971 GFDF01001726 JAV12358.1 GFDF01001727 JAV12357.1 GFDL01008177 JAV26868.1 GBHO01017578 JAG26026.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000005203
UP000036403
+ More
UP000192223 UP000076502 UP000007266 UP000242457 UP000235965 UP000053105 UP000008237 UP000000311 UP000078542 UP000027135 UP000053097 UP000037510 UP000078541 UP000037069 UP000009192 UP000005205 UP000007819 UP000001070 UP000215335 UP000007801 UP000009046 UP000092553 UP000015103 UP000000803 UP000008792 UP000008711 UP000078492 UP000002282 UP000007798 UP000019118 UP000279307 UP000095301 UP000192221 UP000030742 UP000095300 UP000075809 UP000268350 UP000001819 UP000008744 UP000053825 UP000078540 UP000092461 UP000078200 UP000092444 UP000092445 UP000092443 UP000092460
UP000192223 UP000076502 UP000007266 UP000242457 UP000235965 UP000053105 UP000008237 UP000000311 UP000078542 UP000027135 UP000053097 UP000037510 UP000078541 UP000037069 UP000009192 UP000005205 UP000007819 UP000001070 UP000215335 UP000007801 UP000009046 UP000092553 UP000015103 UP000000803 UP000008792 UP000008711 UP000078492 UP000002282 UP000007798 UP000019118 UP000279307 UP000095301 UP000192221 UP000030742 UP000095300 UP000075809 UP000268350 UP000001819 UP000008744 UP000053825 UP000078540 UP000092461 UP000078200 UP000092444 UP000092445 UP000092443 UP000092460
Interpro
Gene 3D
ProteinModelPortal
Q1HPT1
A0A194RTR3
A0A194PE66
I4DM90
K9RZR5
D9ILX6
+ More
Q1HPT0 A0A194RQB3 A0A212EJP1 H9JSP4 I4DMG2 I4DIU5 V9IJZ2 A0A088AMB8 A0A0J7KRQ1 A0A1W4X6X1 A0A154P1L6 D2A4R3 A0A1W6EVQ8 A0A2A3E923 A0A2J7QV00 A0A191UR82 R4WNI7 A0A0N0BJR7 E2C3U0 E2ABW9 A0A2R7VST6 A0A0C9Q3Q8 A0A195BY79 A0A1I9WLI2 A0A067QNJ1 A0A026X0M9 A0A1B6ENI6 V5I8R7 A0A2D1QUC6 A0A0L7KWW5 G9C5G1 A0A195F1S5 A0A0L0BSV2 B4L7D0 A0A158NL77 X1WI47 B4JZN9 A0A232EXM3 A0A023F989 A0A161MK87 Q1ZZQ8 B3N1J9 A0A1L8EHP6 E0W1G3 A0A0M4F807 R4FQF4 L0MQ04 A0A1L8EEU5 D0Z794 X2JH42 B3P9Q0 W0AMY4 Q05825 A0A151JSF9 B4PW41 B4NHL2 J3JX90 A0A3L8DTN4 A0A1I8MLA9 W8BZZ5 T1PAN5 A0A034VA91 A0A1L8EF05 A0A1W4V506 A0A224XET0 B4MEZ1 A0A1P9FG40 U4U6D1 A0A034V7I6 A0A1B6D1T9 A0A1I8NT76 A0A151WHD6 A0A3B0KSV3 Q29CL2 B4H9P4 A0A0A1WKN6 W8BCZ3 A0A0L7RCQ4 U5EUX4 A0A1D1YVH1 A0A1Y1KCU8 A0A195BH40 A0A1L8E9X6 A0A1B0CLT6 A0A1A9V556 D3TRW4 A0A0V0G2S7 A0A1A9ZWH1 A0A1A9YCL3 A0A1B0AQN2 A0A1L8E0V2 A0A1L8E0Y3 A0A1Q3FH39 A0A0A9Y314
Q1HPT0 A0A194RQB3 A0A212EJP1 H9JSP4 I4DMG2 I4DIU5 V9IJZ2 A0A088AMB8 A0A0J7KRQ1 A0A1W4X6X1 A0A154P1L6 D2A4R3 A0A1W6EVQ8 A0A2A3E923 A0A2J7QV00 A0A191UR82 R4WNI7 A0A0N0BJR7 E2C3U0 E2ABW9 A0A2R7VST6 A0A0C9Q3Q8 A0A195BY79 A0A1I9WLI2 A0A067QNJ1 A0A026X0M9 A0A1B6ENI6 V5I8R7 A0A2D1QUC6 A0A0L7KWW5 G9C5G1 A0A195F1S5 A0A0L0BSV2 B4L7D0 A0A158NL77 X1WI47 B4JZN9 A0A232EXM3 A0A023F989 A0A161MK87 Q1ZZQ8 B3N1J9 A0A1L8EHP6 E0W1G3 A0A0M4F807 R4FQF4 L0MQ04 A0A1L8EEU5 D0Z794 X2JH42 B3P9Q0 W0AMY4 Q05825 A0A151JSF9 B4PW41 B4NHL2 J3JX90 A0A3L8DTN4 A0A1I8MLA9 W8BZZ5 T1PAN5 A0A034VA91 A0A1L8EF05 A0A1W4V506 A0A224XET0 B4MEZ1 A0A1P9FG40 U4U6D1 A0A034V7I6 A0A1B6D1T9 A0A1I8NT76 A0A151WHD6 A0A3B0KSV3 Q29CL2 B4H9P4 A0A0A1WKN6 W8BCZ3 A0A0L7RCQ4 U5EUX4 A0A1D1YVH1 A0A1Y1KCU8 A0A195BH40 A0A1L8E9X6 A0A1B0CLT6 A0A1A9V556 D3TRW4 A0A0V0G2S7 A0A1A9ZWH1 A0A1A9YCL3 A0A1B0AQN2 A0A1L8E0V2 A0A1L8E0Y3 A0A1Q3FH39 A0A0A9Y314
PDB
2XND
E-value=0,
Score=1996
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Mitochondrion inner membrane
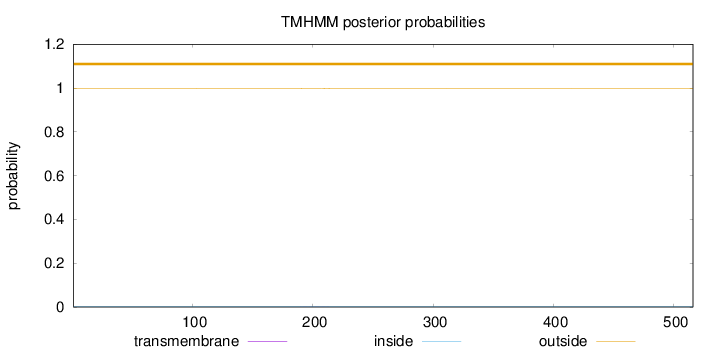
Length:
516
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0246399999999999
Exp number, first 60 AAs:
0.00063
Total prob of N-in:
0.00137
outside
1 - 516
Population Genetic Test Statistics
Pi
149.790405
Theta
133.485894
Tajima's D
0.389252
CLR
0.020229
CSRT
0.48067596620169
Interpretation
Uncertain
