Pre Gene Modal
BGIBMGA012547
Annotation
PREDICTED:_60S_ribosomal_export_protein_NMD3_[Amyelois_transitella]
Full name
60S ribosomal export protein NMD3
Location in the cell
Extracellular Reliability : 1.651 Nuclear Reliability : 1.783
Sequence
CDS
ATGGAATACTTTGCTCCAGAAAACGCTTCTTTAAGCAACAATACACGAATATTGTGCTGTCAGTGTGCTGTACCTATAGAAGCAAACCCCTCAAATATGTGTGTAGCTTGTTTAAGGGCTCATGTAGACATCACAGATGGAATACCCAAACAAGCGACATTATTCTTCTGTAGGGGTTGTGAAAGATATTTGCAACCACCCTCTGAGTGGGTTGTGTGTGCATTAGAGTCACGTGAACTCTTAGCTCTCTGTCTCAAGAGATTAAAAGGATTGAGCAGAGTTAAGTTAATTGATGCTGGTTTTGCTTGGACTGAGCCACATTCGAAGAGAATAAAGGTAAAACTAACTGTACAAGGCGAAGTTATCGGAGGAGCAGTGCTCCAGCAGACATTTATAGTTGAATTTACAATTCAACATCAAATGTGCGATGCTTGCCATCGTTCAGAAGCTCAAGATTACTGGCGTGCTTTAGTGCAAGTACGACAAAGAGCTAATAACAGGAAAACATTTTACTATCTTGAACAATTGATATTGAAGCATAAGGCTCATGAAAATACTCTAGGGATCAAGCCCAAGCATGATGGACTAGATTTCTTCTATTCAACAGAGAATCATGCGCGCAAAATGGTTGATTTTATTCAATCTGTTCTGCCTATAAAATGTCAACATTCCAAGAAACTAATTTCGCATGACATACATAGCAATATATACAACTACAAATTTACATTCAGTGTGGAAATTGTACCTCTATCTAAGGACAGTGTTGTGTGTCTTCCAAAGAAACTTACCCAACAATTGGGATCCATTTCACCAATTTGTTTAGTGCACAGAGTAACTAGCACAATTCATTTGATTGACGCTAATAATGGACAAGGTAAGTTGTGGCCTAAAGAGTCAGACGTCCAATCTACTCATATCGATGTGACAATACTGATGCCCAATTCCTGTAGTCCTATAATATTTTTAACACAAATACATAGCTGA
Protein
MEYFAPENASLSNNTRILCCQCAVPIEANPSNMCVACLRAHVDITDGIPKQATLFFCRGCERYLQPPSEWVVCALESRELLALCLKRLKGLSRVKLIDAGFAWTEPHSKRIKVKLTVQGEVIGGAVLQQTFIVEFTIQHQMCDACHRSEAQDYWRALVQVRQRANNRKTFYYLEQLILKHKAHENTLGIKPKHDGLDFFYSTENHARKMVDFIQSVLPIKCQHSKKLISHDIHSNIYNYKFTFSVEIVPLSKDSVVCLPKKLTQQLGSISPICLVHRVTSTIHLIDANNGQGKLWPKESDVQSTHIDVTILMPNSCSPIIFLTQIHS
Summary
Description
Acts as an adapter for the XPO1/CRM1-mediated export of the 60S ribosomal subunit.
Similarity
Belongs to the NMD3 family.
Uniprot
A0A194RQ27
A0A2H1WIY3
I4DN29
A0A212EJQ2
S4PXW6
A0A194PDA5
+ More
H9JSN6 A0A2A4IT96 A0A2J7RRM1 A0A2J7RRM3 A0A067QU35 A0A0C9R142 A0A0T6B6J3 A0A1Y1KIM4 A0A0C9RHQ5 D6X4U4 A0A0K8TRM1 E2BQM7 E0VQ58 A0A0V0G8L5 A0A0A9Y8T2 A0A224XMR9 K7J8J1 A0A0P4VWJ0 R4FP80 A0A0M8ZSR8 A0A088AJK8 A0A2A3E6R7 A0A2J7RRM9 A0A1B6ET12 A0A1B6I271 A0A2H8TM30 X1WIX7 A0A158NY95 A0A2S2QB92 A0A1I8MNT4 J3JWL7 A0A151ITA0 E9JD41 A0A1W4WGM1 T1PAP7 A0A1A9X037 A0A182SFG9 A0A1I8PHD5 A0A154PDP0 A0A0L7R7D7 A0A182YNN5 A0A336M598 W8BY02 U5EXW8 A0A0L0C4H7 B4JWS1 A0A1B0FCV2 A0A1A9ZLR6 A0A0K8TXD5 A0A0L7KXI1 A0A034VQ24 A0A0A1WFK2 B3MYR9 B4L7V0 A0A1L8DS94 B4MA06 A0A1B6M1Y2 A0A1J1HQZ0 A0A1B6CCI8 A0A0M3QZ98 Q7PNZ3 A0A182HMD5 A0A1C7CZN3 A0A182UNA8 A0A182PB58 A0A182K3T4 A0A182FFP5 O46050 T1D3X6 A0A084WEV6 A0A2M4A964 Q9UB80 W5JC40 B4NQ13 A0A182WAN1 A0A2M3Z4K6 A0A2M3Z471 B4Q196 A0A2M4BKR7 A0A2M4BKX0 A0A182N7U0 A0A182R2N3 Q0IGB3 B3P8X8 Q1HR44 A0A023EUG8 A0A1W4VSF3 B4H380 A0A1Q3G1M9 E9G734 A0A3B0JY72 Q29JL9 A0A195FWB8 A0A151I3Y5
H9JSN6 A0A2A4IT96 A0A2J7RRM1 A0A2J7RRM3 A0A067QU35 A0A0C9R142 A0A0T6B6J3 A0A1Y1KIM4 A0A0C9RHQ5 D6X4U4 A0A0K8TRM1 E2BQM7 E0VQ58 A0A0V0G8L5 A0A0A9Y8T2 A0A224XMR9 K7J8J1 A0A0P4VWJ0 R4FP80 A0A0M8ZSR8 A0A088AJK8 A0A2A3E6R7 A0A2J7RRM9 A0A1B6ET12 A0A1B6I271 A0A2H8TM30 X1WIX7 A0A158NY95 A0A2S2QB92 A0A1I8MNT4 J3JWL7 A0A151ITA0 E9JD41 A0A1W4WGM1 T1PAP7 A0A1A9X037 A0A182SFG9 A0A1I8PHD5 A0A154PDP0 A0A0L7R7D7 A0A182YNN5 A0A336M598 W8BY02 U5EXW8 A0A0L0C4H7 B4JWS1 A0A1B0FCV2 A0A1A9ZLR6 A0A0K8TXD5 A0A0L7KXI1 A0A034VQ24 A0A0A1WFK2 B3MYR9 B4L7V0 A0A1L8DS94 B4MA06 A0A1B6M1Y2 A0A1J1HQZ0 A0A1B6CCI8 A0A0M3QZ98 Q7PNZ3 A0A182HMD5 A0A1C7CZN3 A0A182UNA8 A0A182PB58 A0A182K3T4 A0A182FFP5 O46050 T1D3X6 A0A084WEV6 A0A2M4A964 Q9UB80 W5JC40 B4NQ13 A0A182WAN1 A0A2M3Z4K6 A0A2M3Z471 B4Q196 A0A2M4BKR7 A0A2M4BKX0 A0A182N7U0 A0A182R2N3 Q0IGB3 B3P8X8 Q1HR44 A0A023EUG8 A0A1W4VSF3 B4H380 A0A1Q3G1M9 E9G734 A0A3B0JY72 Q29JL9 A0A195FWB8 A0A151I3Y5
Pubmed
26354079
22651552
22118469
23622113
19121390
24845553
+ More
28004739 18362917 19820115 26369729 20798317 20566863 25401762 26823975 20075255 27129103 21347285 25315136 22516182 23537049 21282665 25244985 24495485 26108605 17994087 26227816 25348373 25830018 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24330624 24438588 20920257 23761445 17550304 17510324 17204158 24945155 26483478 21292972 15632085
28004739 18362917 19820115 26369729 20798317 20566863 25401762 26823975 20075255 27129103 21347285 25315136 22516182 23537049 21282665 25244985 24495485 26108605 17994087 26227816 25348373 25830018 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24330624 24438588 20920257 23761445 17550304 17510324 17204158 24945155 26483478 21292972 15632085
EMBL
KQ459896
KPJ19509.1
ODYU01008886
SOQ52862.1
AK402697
BAM19319.1
+ More
AGBW02014409 OWR41717.1 GAIX01004323 JAA88237.1 KQ459606 KPI91316.1 BABH01032097 BABH01032098 NWSH01008037 PCG62716.1 NEVH01000599 PNF43488.1 PNF43483.1 KK853016 KDR12488.1 GBYB01000586 JAG70353.1 LJIG01009627 KRT82701.1 GEZM01086564 JAV59246.1 GBYB01000583 GBYB01000584 GBYB01000587 GBYB01012732 GBYB01014888 JAG70350.1 JAG70351.1 JAG70354.1 JAG82499.1 JAG84655.1 KQ971381 EEZ97240.1 GDAI01000589 JAI17014.1 GL449784 EFN81992.1 DS235389 EEB15514.1 GECL01001752 JAP04372.1 GBHO01016096 GBRD01008241 GDHC01019812 JAG27508.1 JAG57580.1 JAP98816.1 GFTR01006696 JAW09730.1 GDKW01001532 JAI55063.1 ACPB03014781 GAHY01001014 JAA76496.1 KQ435916 KOX68769.1 KZ288349 PBC27408.1 PNF43486.1 GECZ01028683 JAS41086.1 GECU01026680 JAS81026.1 GFXV01002533 MBW14338.1 ABLF02012664 ABLF02033505 ABLF02047324 ADTU01003507 GGMS01005793 MBY74996.1 BT127635 KB632375 AEE62597.1 ERL93932.1 KQ981033 KYN10172.1 GL771866 EFZ09199.1 KA645779 AFP60408.1 KQ434869 KZC09300.1 KQ414642 KOC66748.1 UFQT01000550 UFQT01002626 SSX25190.1 SSX33791.1 GAMC01005107 JAC01449.1 GANO01000707 JAB59164.1 JRES01000994 KNC26349.1 CH916376 EDV95197.1 CCAG010007386 GDHF01033192 JAI19122.1 JTDY01004796 KOB67746.1 GAKP01015027 GAKP01015024 JAC43925.1 GBXI01016826 JAC97465.1 CH902632 EDV32763.1 CH933814 EDW05525.1 GFDF01004813 JAV09271.1 CH940655 EDW66065.1 GEBQ01010036 JAT29941.1 CVRI01000019 CRK90455.1 GEDC01026125 JAS11173.1 CP012528 ALC48999.1 AAAB01008960 EAA11292.5 APCN01004407 AE014298 AY058494 AL009194 AAF45744.1 AAL13723.1 CAA15695.1 GALA01001183 JAA93669.1 ATLV01023259 KE525341 KFB48750.1 GGFK01003949 MBW37270.1 AJ238705 CAB42049.1 ADMH02001932 ETN60435.1 CH964291 EDW86238.1 GGFM01002682 MBW23433.1 GGFM01002566 MBW23317.1 CM000162 EDX01403.2 GGFJ01004504 MBW53645.1 GGFJ01004503 MBW53644.1 CH477260 EAT45726.1 CH954183 EDV45583.1 DQ440250 ABF18283.1 JXUM01096995 GAPW01001022 KQ564308 JAC12576.1 KXJ72449.1 CH479205 EDW30773.1 GFDL01001331 JAV33714.1 GL732534 EFX84398.1 OUUW01000003 SPP78709.1 CH379063 EAL32282.2 KQ981204 KYN44960.1 KQ976462 KYM84648.1
AGBW02014409 OWR41717.1 GAIX01004323 JAA88237.1 KQ459606 KPI91316.1 BABH01032097 BABH01032098 NWSH01008037 PCG62716.1 NEVH01000599 PNF43488.1 PNF43483.1 KK853016 KDR12488.1 GBYB01000586 JAG70353.1 LJIG01009627 KRT82701.1 GEZM01086564 JAV59246.1 GBYB01000583 GBYB01000584 GBYB01000587 GBYB01012732 GBYB01014888 JAG70350.1 JAG70351.1 JAG70354.1 JAG82499.1 JAG84655.1 KQ971381 EEZ97240.1 GDAI01000589 JAI17014.1 GL449784 EFN81992.1 DS235389 EEB15514.1 GECL01001752 JAP04372.1 GBHO01016096 GBRD01008241 GDHC01019812 JAG27508.1 JAG57580.1 JAP98816.1 GFTR01006696 JAW09730.1 GDKW01001532 JAI55063.1 ACPB03014781 GAHY01001014 JAA76496.1 KQ435916 KOX68769.1 KZ288349 PBC27408.1 PNF43486.1 GECZ01028683 JAS41086.1 GECU01026680 JAS81026.1 GFXV01002533 MBW14338.1 ABLF02012664 ABLF02033505 ABLF02047324 ADTU01003507 GGMS01005793 MBY74996.1 BT127635 KB632375 AEE62597.1 ERL93932.1 KQ981033 KYN10172.1 GL771866 EFZ09199.1 KA645779 AFP60408.1 KQ434869 KZC09300.1 KQ414642 KOC66748.1 UFQT01000550 UFQT01002626 SSX25190.1 SSX33791.1 GAMC01005107 JAC01449.1 GANO01000707 JAB59164.1 JRES01000994 KNC26349.1 CH916376 EDV95197.1 CCAG010007386 GDHF01033192 JAI19122.1 JTDY01004796 KOB67746.1 GAKP01015027 GAKP01015024 JAC43925.1 GBXI01016826 JAC97465.1 CH902632 EDV32763.1 CH933814 EDW05525.1 GFDF01004813 JAV09271.1 CH940655 EDW66065.1 GEBQ01010036 JAT29941.1 CVRI01000019 CRK90455.1 GEDC01026125 JAS11173.1 CP012528 ALC48999.1 AAAB01008960 EAA11292.5 APCN01004407 AE014298 AY058494 AL009194 AAF45744.1 AAL13723.1 CAA15695.1 GALA01001183 JAA93669.1 ATLV01023259 KE525341 KFB48750.1 GGFK01003949 MBW37270.1 AJ238705 CAB42049.1 ADMH02001932 ETN60435.1 CH964291 EDW86238.1 GGFM01002682 MBW23433.1 GGFM01002566 MBW23317.1 CM000162 EDX01403.2 GGFJ01004504 MBW53645.1 GGFJ01004503 MBW53644.1 CH477260 EAT45726.1 CH954183 EDV45583.1 DQ440250 ABF18283.1 JXUM01096995 GAPW01001022 KQ564308 JAC12576.1 KXJ72449.1 CH479205 EDW30773.1 GFDL01001331 JAV33714.1 GL732534 EFX84398.1 OUUW01000003 SPP78709.1 CH379063 EAL32282.2 KQ981204 KYN44960.1 KQ976462 KYM84648.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000005204
UP000218220
UP000235965
+ More
UP000027135 UP000007266 UP000008237 UP000009046 UP000002358 UP000015103 UP000053105 UP000005203 UP000242457 UP000007819 UP000005205 UP000095301 UP000030742 UP000078492 UP000192223 UP000091820 UP000075901 UP000095300 UP000076502 UP000053825 UP000076408 UP000037069 UP000001070 UP000092444 UP000092445 UP000037510 UP000007801 UP000009192 UP000008792 UP000183832 UP000092553 UP000007062 UP000075840 UP000075882 UP000075903 UP000075885 UP000075881 UP000069272 UP000000803 UP000030765 UP000000673 UP000007798 UP000075920 UP000002282 UP000075884 UP000075900 UP000008820 UP000008711 UP000069940 UP000249989 UP000192221 UP000008744 UP000000305 UP000268350 UP000001819 UP000078541 UP000078540
UP000027135 UP000007266 UP000008237 UP000009046 UP000002358 UP000015103 UP000053105 UP000005203 UP000242457 UP000007819 UP000005205 UP000095301 UP000030742 UP000078492 UP000192223 UP000091820 UP000075901 UP000095300 UP000076502 UP000053825 UP000076408 UP000037069 UP000001070 UP000092444 UP000092445 UP000037510 UP000007801 UP000009192 UP000008792 UP000183832 UP000092553 UP000007062 UP000075840 UP000075882 UP000075903 UP000075885 UP000075881 UP000069272 UP000000803 UP000030765 UP000000673 UP000007798 UP000075920 UP000002282 UP000075884 UP000075900 UP000008820 UP000008711 UP000069940 UP000249989 UP000192221 UP000008744 UP000000305 UP000268350 UP000001819 UP000078541 UP000078540
Interpro
SUPFAM
SSF47862
SSF47862
ProteinModelPortal
A0A194RQ27
A0A2H1WIY3
I4DN29
A0A212EJQ2
S4PXW6
A0A194PDA5
+ More
H9JSN6 A0A2A4IT96 A0A2J7RRM1 A0A2J7RRM3 A0A067QU35 A0A0C9R142 A0A0T6B6J3 A0A1Y1KIM4 A0A0C9RHQ5 D6X4U4 A0A0K8TRM1 E2BQM7 E0VQ58 A0A0V0G8L5 A0A0A9Y8T2 A0A224XMR9 K7J8J1 A0A0P4VWJ0 R4FP80 A0A0M8ZSR8 A0A088AJK8 A0A2A3E6R7 A0A2J7RRM9 A0A1B6ET12 A0A1B6I271 A0A2H8TM30 X1WIX7 A0A158NY95 A0A2S2QB92 A0A1I8MNT4 J3JWL7 A0A151ITA0 E9JD41 A0A1W4WGM1 T1PAP7 A0A1A9X037 A0A182SFG9 A0A1I8PHD5 A0A154PDP0 A0A0L7R7D7 A0A182YNN5 A0A336M598 W8BY02 U5EXW8 A0A0L0C4H7 B4JWS1 A0A1B0FCV2 A0A1A9ZLR6 A0A0K8TXD5 A0A0L7KXI1 A0A034VQ24 A0A0A1WFK2 B3MYR9 B4L7V0 A0A1L8DS94 B4MA06 A0A1B6M1Y2 A0A1J1HQZ0 A0A1B6CCI8 A0A0M3QZ98 Q7PNZ3 A0A182HMD5 A0A1C7CZN3 A0A182UNA8 A0A182PB58 A0A182K3T4 A0A182FFP5 O46050 T1D3X6 A0A084WEV6 A0A2M4A964 Q9UB80 W5JC40 B4NQ13 A0A182WAN1 A0A2M3Z4K6 A0A2M3Z471 B4Q196 A0A2M4BKR7 A0A2M4BKX0 A0A182N7U0 A0A182R2N3 Q0IGB3 B3P8X8 Q1HR44 A0A023EUG8 A0A1W4VSF3 B4H380 A0A1Q3G1M9 E9G734 A0A3B0JY72 Q29JL9 A0A195FWB8 A0A151I3Y5
H9JSN6 A0A2A4IT96 A0A2J7RRM1 A0A2J7RRM3 A0A067QU35 A0A0C9R142 A0A0T6B6J3 A0A1Y1KIM4 A0A0C9RHQ5 D6X4U4 A0A0K8TRM1 E2BQM7 E0VQ58 A0A0V0G8L5 A0A0A9Y8T2 A0A224XMR9 K7J8J1 A0A0P4VWJ0 R4FP80 A0A0M8ZSR8 A0A088AJK8 A0A2A3E6R7 A0A2J7RRM9 A0A1B6ET12 A0A1B6I271 A0A2H8TM30 X1WIX7 A0A158NY95 A0A2S2QB92 A0A1I8MNT4 J3JWL7 A0A151ITA0 E9JD41 A0A1W4WGM1 T1PAP7 A0A1A9X037 A0A182SFG9 A0A1I8PHD5 A0A154PDP0 A0A0L7R7D7 A0A182YNN5 A0A336M598 W8BY02 U5EXW8 A0A0L0C4H7 B4JWS1 A0A1B0FCV2 A0A1A9ZLR6 A0A0K8TXD5 A0A0L7KXI1 A0A034VQ24 A0A0A1WFK2 B3MYR9 B4L7V0 A0A1L8DS94 B4MA06 A0A1B6M1Y2 A0A1J1HQZ0 A0A1B6CCI8 A0A0M3QZ98 Q7PNZ3 A0A182HMD5 A0A1C7CZN3 A0A182UNA8 A0A182PB58 A0A182K3T4 A0A182FFP5 O46050 T1D3X6 A0A084WEV6 A0A2M4A964 Q9UB80 W5JC40 B4NQ13 A0A182WAN1 A0A2M3Z4K6 A0A2M3Z471 B4Q196 A0A2M4BKR7 A0A2M4BKX0 A0A182N7U0 A0A182R2N3 Q0IGB3 B3P8X8 Q1HR44 A0A023EUG8 A0A1W4VSF3 B4H380 A0A1Q3G1M9 E9G734 A0A3B0JY72 Q29JL9 A0A195FWB8 A0A151I3Y5
PDB
5T6R
E-value=2.67868e-82,
Score=777
Ontologies
KEGG
PATHWAY
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
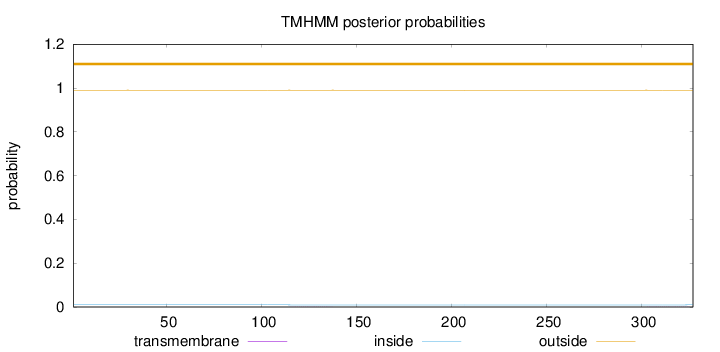
Length:
327
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01186
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00974
outside
1 - 327
Population Genetic Test Statistics
Pi
244.105955
Theta
193.860347
Tajima's D
0.867678
CLR
0
CSRT
0.623018849057547
Interpretation
Possibly Positive selection
