Gene
KWMTBOMO05066 Validated by peptides from experiments
Annotation
UPF0389_protein_[Operophtera_brumata]
Full name
UPF0389 protein CG9231
+ More
UPF0389 protein GA21628
UPF0389 protein GA21628
Location in the cell
Nuclear Reliability : 2.415
Sequence
CDS
ATGTGTTCATCTTCACCGACAGGACCCAGTTCAACAGGTCCATCATCATCATCTATGTCTCATAGATTTCGACCGACAGAATTTCAGAAAACGATTCTTGTGTGGACAAAAAAATACAAGAACAAATCTGAAGTCCCTCCATTTGTATCAGCTGAAATTATTGAAAGATCTAAGAGTGAAGCTAGAATCAAAATTTCAAATGTGCTTATGCTTCTTACTGCTTTGGCAAGTTTTGGAGCAATATTGTCTGGGAAAGCTGCTGCTAAGAGAGGGGAATCTGTTCATCAGATGAATCTTGACTGGCATAAAAAATATCAAGAAGAACATAAAGAAAAAAGTAGTGCAAAGTAA
Protein
MCSSSPTGPSSTGPSSSSMSHRFRPTEFQKTILVWTKKYKNKSEVPPFVSAEIIERSKSEARIKISNVLMLLTALASFGAILSGKAAAKRGESVHQMNLDWHKKYQEEHKEKSSAK
Summary
Similarity
Belongs to the UPF0389 family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain UPF0389 protein CG9231
Uniprot
A0A0L7KWY3
A0A194RNU9
A0A194PDI5
A0A3S2LHP6
A0A2H1WUQ7
A0A1E1WEG4
+ More
S4P9R1 A0A212EJS7 A0A1J1HVG6 B0WAK7 A0A023EHP4 Q1HQM7 A0A1B0C0B5 A0A1L8E9D3 A0A1L8E9N8 A0A1L8E9R4 A0A023EHN7 A0A1W7R801 A0A1Q3FG97 A0A1Q3FGQ2 B4QQV2 A0A1B0GD62 B3NIF4 A0A1A9ZS69 X2JGV8 Q9VW12 A0A1W4URC7 B4IIM5 A0A0V0G9S3 A0A182JP53 A0A034WN90 A0A1B0CUG5 A0A1A9UQI2 A0A0K8V4J6 A0A0N7Z8E8 B4IUC1 A0A182SIR1 T1IG06 A0A182NK36 A0A2M4AU23 A0A0C9R0Q1 B3M700 A0A182PCT5 A0A182RDM4 T1DR25 W5JSC6 A0A182QDN9 A0A023FBJ9 A0A182YKT1 A0A1I8MM20 A0A182MVE3 A0A182FH59 A0A1C7CZ60 A0A182I2L0 A0A182WX15 A0A182VEZ1 Q7QIZ6 A0A182TDG0 A0A2M3ZIX0 A0A2M3ZB33 A0A2M4C221 A0A2M3ZCS0 A0A2M3ZIW7 A0A182WLK6 Q29DG0 D6WCW9 A0A1L8DUT7 B4H998 A0A1L8DUS0 A0A3B0J9I4 A0A1L8DUT2 A0A0A1WQK9 A0A182J8P1 A0A084WEM3 B4LCL9 V5I942 B4KZC5 A0A0M4EQX9 A0A1B6GAP2 A0A336JY21 A0A336LHZ6 A0A0P5DM24 A0A0P4WFB8 A0A1L8DUS6 K7J0G2 B4MXL7 A0A232F410 A0A0P6FED8 A0A0P4W9V1 A0A0A9X4I1 B4J1C2 A0A1L8DUR3 A0A0N8CBD7 E9GIP3 A0A0P6GUI1 A0A0P5YUW8 A0A0P6BUQ8 A0A1Y1L502 E9GIP2
S4P9R1 A0A212EJS7 A0A1J1HVG6 B0WAK7 A0A023EHP4 Q1HQM7 A0A1B0C0B5 A0A1L8E9D3 A0A1L8E9N8 A0A1L8E9R4 A0A023EHN7 A0A1W7R801 A0A1Q3FG97 A0A1Q3FGQ2 B4QQV2 A0A1B0GD62 B3NIF4 A0A1A9ZS69 X2JGV8 Q9VW12 A0A1W4URC7 B4IIM5 A0A0V0G9S3 A0A182JP53 A0A034WN90 A0A1B0CUG5 A0A1A9UQI2 A0A0K8V4J6 A0A0N7Z8E8 B4IUC1 A0A182SIR1 T1IG06 A0A182NK36 A0A2M4AU23 A0A0C9R0Q1 B3M700 A0A182PCT5 A0A182RDM4 T1DR25 W5JSC6 A0A182QDN9 A0A023FBJ9 A0A182YKT1 A0A1I8MM20 A0A182MVE3 A0A182FH59 A0A1C7CZ60 A0A182I2L0 A0A182WX15 A0A182VEZ1 Q7QIZ6 A0A182TDG0 A0A2M3ZIX0 A0A2M3ZB33 A0A2M4C221 A0A2M3ZCS0 A0A2M3ZIW7 A0A182WLK6 Q29DG0 D6WCW9 A0A1L8DUT7 B4H998 A0A1L8DUS0 A0A3B0J9I4 A0A1L8DUT2 A0A0A1WQK9 A0A182J8P1 A0A084WEM3 B4LCL9 V5I942 B4KZC5 A0A0M4EQX9 A0A1B6GAP2 A0A336JY21 A0A336LHZ6 A0A0P5DM24 A0A0P4WFB8 A0A1L8DUS6 K7J0G2 B4MXL7 A0A232F410 A0A0P6FED8 A0A0P4W9V1 A0A0A9X4I1 B4J1C2 A0A1L8DUR3 A0A0N8CBD7 E9GIP3 A0A0P6GUI1 A0A0P5YUW8 A0A0P6BUQ8 A0A1Y1L502 E9GIP2
Pubmed
26227816
26354079
23622113
22118469
24945155
17204158
+ More
17510324 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17893096 25348373 27129103 17550304 20920257 23761445 25474469 25244985 25315136 20966253 12364791 14747013 17210077 15632085 18362917 19820115 25830018 24438588 20075255 28648823 25401762 26823975 21292972 28004739
17510324 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17893096 25348373 27129103 17550304 20920257 23761445 25474469 25244985 25315136 20966253 12364791 14747013 17210077 15632085 18362917 19820115 25830018 24438588 20075255 28648823 25401762 26823975 21292972 28004739
EMBL
JTDY01004796
KOB67747.1
KQ459896
KPJ19513.1
KQ459606
KPI91312.1
+ More
RSAL01000007 RVE54050.1 ODYU01011211 SOQ56809.1 GDQN01005737 JAT85317.1 GAIX01003639 JAA88921.1 AGBW02014409 OWR41721.1 CVRI01000020 CRK91348.1 DS231873 EDS41561.1 GAPW01005197 JAC08401.1 DQ440417 CH477239 ABF18450.1 EAT46495.1 JXJN01023510 GFDG01003495 JAV15304.1 GFDG01003494 JAV15305.1 GFDG01003496 JAV15303.1 GAPW01005198 JAC08400.1 GEHC01000371 JAV47274.1 GFDL01008469 JAV26576.1 GFDL01008330 JAV26715.1 CM000363 CM002912 EDX11090.1 KMZ00565.1 CCAG010002320 CH954178 EDV52379.1 AE014296 AHN58145.1 AY094812 CH480844 EDW49796.1 GECL01001329 JAP04795.1 GAKP01001901 JAC57051.1 AJWK01029162 GDHF01018500 GDHF01017302 JAI33814.1 JAI35012.1 GDKW01003547 JAI53048.1 CM000159 CH891846 EDW95311.1 EDW99984.1 ACPB03011086 GGFK01010943 MBW44264.1 GBYB01006497 JAG76264.1 CH902618 EDV40865.1 GAMD01000840 JAB00751.1 ADMH02000310 ETN67021.1 AXCN02000433 GBBI01000414 JAC18298.1 AXCM01004731 APCN01000169 AAAB01008807 EAA04250.3 EDO64694.1 GGFM01007730 MBW28481.1 GGFM01004929 MBW25680.1 GGFJ01010173 MBW59314.1 GGFM01005499 MBW26250.1 GGFM01007702 MBW28453.1 CH379070 KQ971311 EEZ98829.1 GFDF01003898 JAV10186.1 CH479227 EDW35322.1 GFDF01003897 JAV10187.1 OUUW01000002 SPP76602.1 GFDF01003905 JAV10179.1 GBXI01013361 JAD00931.1 ATLV01023228 KE525341 KFB48667.1 CH940647 EDW69882.1 GALX01003590 JAB64876.1 CH933809 EDW18951.1 CP012525 ALC44767.1 GECZ01010268 JAS59501.1 UFQS01000007 UFQT01000007 SSW97121.1 SSX17507.1 SSW97120.1 SSX17506.1 GDIP01154386 LRGB01000568 JAJ69016.1 KZS18150.1 GDRN01061424 JAI65210.1 GFDF01003894 JAV10190.1 CH963876 EDW76786.1 NNAY01001049 OXU25342.1 GDIQ01192122 GDIQ01048887 JAN45850.1 KZS18148.1 GDRN01061425 JAI65209.1 GBHO01029910 GBHO01029909 GBHO01029907 GBHO01029906 GBRD01015554 GBRD01015553 GDHC01013733 GDHC01000020 JAG13694.1 JAG13695.1 JAG13697.1 JAG13698.1 JAG50272.1 JAQ04896.1 JAQ18609.1 CH916366 EDV97991.1 GFDF01003900 JAV10184.1 GDIQ01096363 JAL55363.1 GL732546 EFX80691.1 GDIQ01029143 JAN65594.1 GDIP01053076 JAM50639.1 GDIP01009550 JAM94165.1 GEZM01064505 JAV68753.1 EFX80690.1
RSAL01000007 RVE54050.1 ODYU01011211 SOQ56809.1 GDQN01005737 JAT85317.1 GAIX01003639 JAA88921.1 AGBW02014409 OWR41721.1 CVRI01000020 CRK91348.1 DS231873 EDS41561.1 GAPW01005197 JAC08401.1 DQ440417 CH477239 ABF18450.1 EAT46495.1 JXJN01023510 GFDG01003495 JAV15304.1 GFDG01003494 JAV15305.1 GFDG01003496 JAV15303.1 GAPW01005198 JAC08400.1 GEHC01000371 JAV47274.1 GFDL01008469 JAV26576.1 GFDL01008330 JAV26715.1 CM000363 CM002912 EDX11090.1 KMZ00565.1 CCAG010002320 CH954178 EDV52379.1 AE014296 AHN58145.1 AY094812 CH480844 EDW49796.1 GECL01001329 JAP04795.1 GAKP01001901 JAC57051.1 AJWK01029162 GDHF01018500 GDHF01017302 JAI33814.1 JAI35012.1 GDKW01003547 JAI53048.1 CM000159 CH891846 EDW95311.1 EDW99984.1 ACPB03011086 GGFK01010943 MBW44264.1 GBYB01006497 JAG76264.1 CH902618 EDV40865.1 GAMD01000840 JAB00751.1 ADMH02000310 ETN67021.1 AXCN02000433 GBBI01000414 JAC18298.1 AXCM01004731 APCN01000169 AAAB01008807 EAA04250.3 EDO64694.1 GGFM01007730 MBW28481.1 GGFM01004929 MBW25680.1 GGFJ01010173 MBW59314.1 GGFM01005499 MBW26250.1 GGFM01007702 MBW28453.1 CH379070 KQ971311 EEZ98829.1 GFDF01003898 JAV10186.1 CH479227 EDW35322.1 GFDF01003897 JAV10187.1 OUUW01000002 SPP76602.1 GFDF01003905 JAV10179.1 GBXI01013361 JAD00931.1 ATLV01023228 KE525341 KFB48667.1 CH940647 EDW69882.1 GALX01003590 JAB64876.1 CH933809 EDW18951.1 CP012525 ALC44767.1 GECZ01010268 JAS59501.1 UFQS01000007 UFQT01000007 SSW97121.1 SSX17507.1 SSW97120.1 SSX17506.1 GDIP01154386 LRGB01000568 JAJ69016.1 KZS18150.1 GDRN01061424 JAI65210.1 GFDF01003894 JAV10190.1 CH963876 EDW76786.1 NNAY01001049 OXU25342.1 GDIQ01192122 GDIQ01048887 JAN45850.1 KZS18148.1 GDRN01061425 JAI65209.1 GBHO01029910 GBHO01029909 GBHO01029907 GBHO01029906 GBRD01015554 GBRD01015553 GDHC01013733 GDHC01000020 JAG13694.1 JAG13695.1 JAG13697.1 JAG13698.1 JAG50272.1 JAQ04896.1 JAQ18609.1 CH916366 EDV97991.1 GFDF01003900 JAV10184.1 GDIQ01096363 JAL55363.1 GL732546 EFX80691.1 GDIQ01029143 JAN65594.1 GDIP01053076 JAM50639.1 GDIP01009550 JAM94165.1 GEZM01064505 JAV68753.1 EFX80690.1
Proteomes
UP000037510
UP000053240
UP000053268
UP000283053
UP000007151
UP000183832
+ More
UP000002320 UP000008820 UP000092460 UP000000304 UP000092444 UP000008711 UP000092445 UP000000803 UP000192221 UP000001292 UP000075881 UP000092461 UP000078200 UP000002282 UP000075901 UP000015103 UP000075884 UP000007801 UP000075885 UP000075900 UP000000673 UP000075886 UP000076408 UP000095301 UP000075883 UP000069272 UP000075882 UP000075840 UP000076407 UP000075903 UP000007062 UP000075902 UP000075920 UP000001819 UP000007266 UP000008744 UP000268350 UP000075880 UP000030765 UP000008792 UP000009192 UP000092553 UP000076858 UP000002358 UP000007798 UP000215335 UP000001070 UP000000305
UP000002320 UP000008820 UP000092460 UP000000304 UP000092444 UP000008711 UP000092445 UP000000803 UP000192221 UP000001292 UP000075881 UP000092461 UP000078200 UP000002282 UP000075901 UP000015103 UP000075884 UP000007801 UP000075885 UP000075900 UP000000673 UP000075886 UP000076408 UP000095301 UP000075883 UP000069272 UP000075882 UP000075840 UP000076407 UP000075903 UP000007062 UP000075902 UP000075920 UP000001819 UP000007266 UP000008744 UP000268350 UP000075880 UP000030765 UP000008792 UP000009192 UP000092553 UP000076858 UP000002358 UP000007798 UP000215335 UP000001070 UP000000305
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
ProteinModelPortal
A0A0L7KWY3
A0A194RNU9
A0A194PDI5
A0A3S2LHP6
A0A2H1WUQ7
A0A1E1WEG4
+ More
S4P9R1 A0A212EJS7 A0A1J1HVG6 B0WAK7 A0A023EHP4 Q1HQM7 A0A1B0C0B5 A0A1L8E9D3 A0A1L8E9N8 A0A1L8E9R4 A0A023EHN7 A0A1W7R801 A0A1Q3FG97 A0A1Q3FGQ2 B4QQV2 A0A1B0GD62 B3NIF4 A0A1A9ZS69 X2JGV8 Q9VW12 A0A1W4URC7 B4IIM5 A0A0V0G9S3 A0A182JP53 A0A034WN90 A0A1B0CUG5 A0A1A9UQI2 A0A0K8V4J6 A0A0N7Z8E8 B4IUC1 A0A182SIR1 T1IG06 A0A182NK36 A0A2M4AU23 A0A0C9R0Q1 B3M700 A0A182PCT5 A0A182RDM4 T1DR25 W5JSC6 A0A182QDN9 A0A023FBJ9 A0A182YKT1 A0A1I8MM20 A0A182MVE3 A0A182FH59 A0A1C7CZ60 A0A182I2L0 A0A182WX15 A0A182VEZ1 Q7QIZ6 A0A182TDG0 A0A2M3ZIX0 A0A2M3ZB33 A0A2M4C221 A0A2M3ZCS0 A0A2M3ZIW7 A0A182WLK6 Q29DG0 D6WCW9 A0A1L8DUT7 B4H998 A0A1L8DUS0 A0A3B0J9I4 A0A1L8DUT2 A0A0A1WQK9 A0A182J8P1 A0A084WEM3 B4LCL9 V5I942 B4KZC5 A0A0M4EQX9 A0A1B6GAP2 A0A336JY21 A0A336LHZ6 A0A0P5DM24 A0A0P4WFB8 A0A1L8DUS6 K7J0G2 B4MXL7 A0A232F410 A0A0P6FED8 A0A0P4W9V1 A0A0A9X4I1 B4J1C2 A0A1L8DUR3 A0A0N8CBD7 E9GIP3 A0A0P6GUI1 A0A0P5YUW8 A0A0P6BUQ8 A0A1Y1L502 E9GIP2
S4P9R1 A0A212EJS7 A0A1J1HVG6 B0WAK7 A0A023EHP4 Q1HQM7 A0A1B0C0B5 A0A1L8E9D3 A0A1L8E9N8 A0A1L8E9R4 A0A023EHN7 A0A1W7R801 A0A1Q3FG97 A0A1Q3FGQ2 B4QQV2 A0A1B0GD62 B3NIF4 A0A1A9ZS69 X2JGV8 Q9VW12 A0A1W4URC7 B4IIM5 A0A0V0G9S3 A0A182JP53 A0A034WN90 A0A1B0CUG5 A0A1A9UQI2 A0A0K8V4J6 A0A0N7Z8E8 B4IUC1 A0A182SIR1 T1IG06 A0A182NK36 A0A2M4AU23 A0A0C9R0Q1 B3M700 A0A182PCT5 A0A182RDM4 T1DR25 W5JSC6 A0A182QDN9 A0A023FBJ9 A0A182YKT1 A0A1I8MM20 A0A182MVE3 A0A182FH59 A0A1C7CZ60 A0A182I2L0 A0A182WX15 A0A182VEZ1 Q7QIZ6 A0A182TDG0 A0A2M3ZIX0 A0A2M3ZB33 A0A2M4C221 A0A2M3ZCS0 A0A2M3ZIW7 A0A182WLK6 Q29DG0 D6WCW9 A0A1L8DUT7 B4H998 A0A1L8DUS0 A0A3B0J9I4 A0A1L8DUT2 A0A0A1WQK9 A0A182J8P1 A0A084WEM3 B4LCL9 V5I942 B4KZC5 A0A0M4EQX9 A0A1B6GAP2 A0A336JY21 A0A336LHZ6 A0A0P5DM24 A0A0P4WFB8 A0A1L8DUS6 K7J0G2 B4MXL7 A0A232F410 A0A0P6FED8 A0A0P4W9V1 A0A0A9X4I1 B4J1C2 A0A1L8DUR3 A0A0N8CBD7 E9GIP3 A0A0P6GUI1 A0A0P5YUW8 A0A0P6BUQ8 A0A1Y1L502 E9GIP2
Ontologies
PANTHER
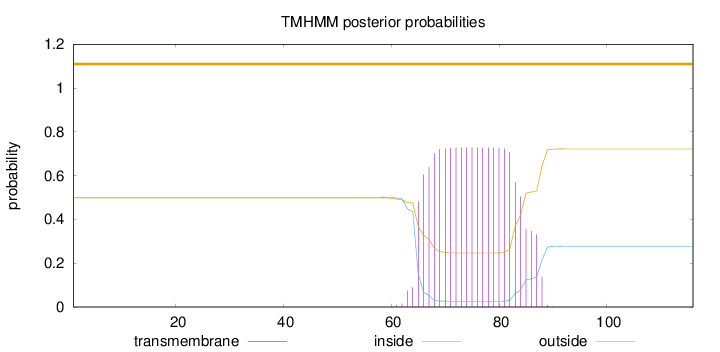
Topology
Subcellular location
Membrane
Length:
116
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.02737
Exp number, first 60 AAs:
0.0026
Total prob of N-in:
0.50164
outside
1 - 116
Population Genetic Test Statistics
Pi
1.471663
Theta
6.954737
Tajima's D
-2.279167
CLR
10.289564
CSRT
0.00224988750562472
Interpretation
Uncertain
