Gene
KWMTBOMO05055 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012560
Annotation
glycerol-3-phosphate_dehydrogenase-1_[Bombyx_mori]
Full name
Glycerol-3-phosphate dehydrogenase [NAD(+)]
+ More
Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic
Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic
Location in the cell
Cytoplasmic Reliability : 3.084
Sequence
CDS
ATGGCAGATAAACAGCCAAAGAACAAAGTTTGCATCGTTGGATCGGGAAACTGGGGTTCTGCTATTGCGAAAATTGTTGGTCGAAATGCTGCGAGTCTATCAAATTTTGAGGATAGAGTTACAATGTGGGTGTATGAAGAAATAATTGAAGGAAAGAAGTTAACTGAAATAATCAATGAAACTCATGAAAATGTTAAATACCTGCCTGGCCACAAATTGCCTTCTAATGTTGTTGCTGTTCCAGATGTAGTTGAAGCTGCAAAAGATGCTGATCTTTTAATATTTGTGGTGCCTCATCAATTTGTCAGAACTATCTGCTCTACTTTGCTTGGAAAAATAAAGCCAACTGCAGCTGCTCTGTCTTTGATTAAGGGATTTGATATAGCCGAAGGTGGTGGCATCGATCTTATATCACATATTATTACAAGATGCCTAAAAATTCCCTGTGCTGTATTAATGGGAGCCAATATTGCATCGGAGGTTGCTGAGGAAAAATTCTGCGAAACGACCATTGGTTGTCGGGACGTAATGCTGGCTCCGTTAATGCGGGATATCATACAGACAGACTACTTCAGGGTCGTGGTGGTGGACGATGAGGACGCAGTCGAAATATGTGGAGCGTTAAAGAACATTGTGGCAGTGGGTGCGGGTTTCGTGGATGGCCTCGGCTACGGTGATAACACAAAGGCTGCTGTCATCAGACTCGGTCTCATGGAGATGATCAAATTCGTTGATGTGTTCTACCCGGGAAGTAAGCTGAGCACGTTCTTCGAGTCATGCGGTGTCGCAGATTTAATAACGACCTGCTACGGGGGCAGAAACAGACGAGTTGCCGAAGCGTTTGTTAAAACGGGACGGTCAATAAAAGAGCTGGAAGACGAAATGCTTAACGGTCAGAAACTACAAGGTCCTATCACCGCGGAGGAAGTAAACCATATGCTCGCTAATAAAAATATGGAAAACAAGTTCCCTTTGTTCACTGCGGTGTTCCGCATCTGTCGCGGAGAACTCAAACCGAACGACTTTATTGATTGCATTCGCAGTCACCCGGAACACATGAAACTGCCCTTGGTCAAATGTTCACTGTGA
Protein
MADKQPKNKVCIVGSGNWGSAIAKIVGRNAASLSNFEDRVTMWVYEEIIEGKKLTEIINETHENVKYLPGHKLPSNVVAVPDVVEAAKDADLLIFVVPHQFVRTICSTLLGKIKPTAAALSLIKGFDIAEGGGIDLISHIITRCLKIPCAVLMGANIASEVAEEKFCETTIGCRDVMLAPLMRDIIQTDYFRVVVVDDEDAVEICGALKNIVAVGAGFVDGLGYGDNTKAAVIRLGLMEMIKFVDVFYPGSKLSTFFESCGVADLITTCYGGRNRRVAEAFVKTGRSIKELEDEMLNGQKLQGPITAEEVNHMLANKNMENKFPLFTAVFRICRGELKPNDFIDCIRSHPEHMKLPLVKCSL
Summary
Catalytic Activity
NAD(+) + sn-glycerol 3-phosphate = dihydroxyacetone phosphate + H(+) + NADH
Subunit
Homodimer.
Similarity
Belongs to the NAD-dependent glycerol-3-phosphate dehydrogenase family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the MT-A70-like family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the MT-A70-like family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Direct protein sequencing
NAD
Oxidoreductase
Polymorphism
Reference proteome
Feature
chain Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic
splice variant In isoform GPDH-3.
splice variant In isoform GPDH-3.
Uniprot
Q68AX4
Q68AX3
H9JSP9
A0A2D3E1P0
A0A194PDH5
A0A0N1PI50
+ More
A0A212FLJ1 A0A2A4JAM7 A0A2A4JBU8 U3MGC9 A0A2H1W0H9 A0A0K8TP17 A0A182XZ58 A0A139WMI0 A0A139WMM5 D6W8W3 V5GW65 T1E8J2 A0A2M4CUC1 A0A2M3Z2N7 A0A2M4A4L1 A0A2M4A4J8 W5JX93 A0A182FI57 A0A182QY36 A0A182IJN4 A0A182RHS9 Q17KS5 A0A1S4EZ75 A0A182P3N4 A0A2M3Z2P5 N6TZN6 A0A1L8E105 Q17KS3 A0A084VWX8 A0A1Y1M526 A0A1J1J5C4 A0A1B0CDX8 J3JZC8 A0A0M9A1K4 Q17KS4 A0A182GGV8 U5EKF5 E2B565 A0A182WIL2 A0A023ERI3 U4UBB4 K7IYD0 A0A232F8R0 A7UR88 A0A182I3L1 A0A182V4T7 Q7PTH2 A0A1Q3FKV5 A0A182KRQ9 B0VZW3 A0A182X3F7 A0A158NXX9 A0A182MJ04 A0A0L0CEP4 A0A0T6BB32 A0A1Y9H2M9 A0A1I8Q1S4 A0A1I8Q1T4 Q6T312 A0A1I8Q1V2 E2AP99 W8BB57 B4MV07 A0A1L8EGZ8 A0A1L8E884 A0A026X5E1 W8AMA6 T1E2R4 A0A1I8Q1V3 T1PP02 A0A1B6CMA5 T1PLW5 P13706-2 A0A0J9QXF0 M9PC43 A0A0R1DLB1 T1PD78 P13706 P13706-4 P13706-3 M9PET0 B4NZU0 A0A0N8NZ96 B4Q441 B5RIM9 B3MMP0 A0A3B0JVE2 A0A0Q5W9H8 B4JBT5 B3N590 Q27928-3 O97463-2 A0A1W4UNC2 Q9TYF7
A0A212FLJ1 A0A2A4JAM7 A0A2A4JBU8 U3MGC9 A0A2H1W0H9 A0A0K8TP17 A0A182XZ58 A0A139WMI0 A0A139WMM5 D6W8W3 V5GW65 T1E8J2 A0A2M4CUC1 A0A2M3Z2N7 A0A2M4A4L1 A0A2M4A4J8 W5JX93 A0A182FI57 A0A182QY36 A0A182IJN4 A0A182RHS9 Q17KS5 A0A1S4EZ75 A0A182P3N4 A0A2M3Z2P5 N6TZN6 A0A1L8E105 Q17KS3 A0A084VWX8 A0A1Y1M526 A0A1J1J5C4 A0A1B0CDX8 J3JZC8 A0A0M9A1K4 Q17KS4 A0A182GGV8 U5EKF5 E2B565 A0A182WIL2 A0A023ERI3 U4UBB4 K7IYD0 A0A232F8R0 A7UR88 A0A182I3L1 A0A182V4T7 Q7PTH2 A0A1Q3FKV5 A0A182KRQ9 B0VZW3 A0A182X3F7 A0A158NXX9 A0A182MJ04 A0A0L0CEP4 A0A0T6BB32 A0A1Y9H2M9 A0A1I8Q1S4 A0A1I8Q1T4 Q6T312 A0A1I8Q1V2 E2AP99 W8BB57 B4MV07 A0A1L8EGZ8 A0A1L8E884 A0A026X5E1 W8AMA6 T1E2R4 A0A1I8Q1V3 T1PP02 A0A1B6CMA5 T1PLW5 P13706-2 A0A0J9QXF0 M9PC43 A0A0R1DLB1 T1PD78 P13706 P13706-4 P13706-3 M9PET0 B4NZU0 A0A0N8NZ96 B4Q441 B5RIM9 B3MMP0 A0A3B0JVE2 A0A0Q5W9H8 B4JBT5 B3N590 Q27928-3 O97463-2 A0A1W4UNC2 Q9TYF7
EC Number
1.1.1.8
Pubmed
19121390
26354079
22118469
23948473
26369729
25244985
+ More
18362917 19820115 20920257 23761445 17510324 23537049 24438588 28004739 22516182 26483478 20798317 24945155 20075255 28648823 12364791 14747013 17210077 20966253 21347285 26108605 24495485 17994087 24508170 30249741 24330624 25315136 2500660 2511555 8094564 7633467 10731132 12537572 12537569 7993372 3755720 2839508 22936249 12537568 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356 8587133 15632085 15545653 9010132 9007023 8522168
18362917 19820115 20920257 23761445 17510324 23537049 24438588 28004739 22516182 26483478 20798317 24945155 20075255 28648823 12364791 14747013 17210077 20966253 21347285 26108605 24495485 17994087 24508170 30249741 24330624 25315136 2500660 2511555 8094564 7633467 10731132 12537572 12537569 7993372 3755720 2839508 22936249 12537568 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356 8587133 15632085 15545653 9010132 9007023 8522168
EMBL
AB164060
BAD38674.1
AB164061
BAD38675.1
BABH01032141
KY565577
+ More
ATU31389.1 KQ459606 KPI91302.1 KQ461133 KPJ08866.1 AGBW02007748 OWR54615.1 NWSH01002106 PCG69167.1 PCG69168.1 KF170736 AGW15345.1 ODYU01005592 SOQ46581.1 GDAI01001486 JAI16117.1 KQ971312 KYB29132.1 KYB29133.1 EEZ98249.2 GALX01004003 JAB64463.1 GAMD01002723 JAA98867.1 GGFL01004735 MBW68913.1 GGFM01002002 MBW22753.1 GGFK01002416 MBW35737.1 GGFK01002415 MBW35736.1 ADMH02000065 ETN67939.1 AXCN02000594 CH477221 EAT47333.1 GGFM01001967 MBW22718.1 APGK01048501 KB741112 ENN73826.1 GFDF01001832 JAV12252.1 EAT47334.1 ATLV01017808 KE525192 KFB42472.1 GEZM01044288 JAV78327.1 CVRI01000073 CRL07613.1 AJWK01008357 AJWK01008358 BT128609 AEE63566.1 KQ435762 KOX75460.1 EAT47332.1 JXUM01011468 JXUM01011469 JXUM01011470 JXUM01011471 KQ560331 KXJ83002.1 GANO01001833 JAB58038.1 GL445725 EFN89159.1 GAPW01002322 JAC11276.1 KB632184 ERL89623.1 NNAY01000716 OXU26833.1 AAAB01008807 EDO64530.1 APCN01000215 EAA03917.3 GFDL01006826 JAV28219.1 DS231815 EDS35205.1 ADTU01003700 AXCM01000512 JRES01000501 KNC30725.1 LJIG01002821 KRT84119.1 AY438978 AAR13229.1 GL441497 EFN64733.1 GAMC01019481 JAB87074.1 CH963857 EDW76352.1 GFDG01000838 JAV17961.1 GFDG01003888 JAV14911.1 KK107019 QOIP01000007 EZA62634.1 RLU20422.1 GAMC01019478 JAB87077.1 GALA01001060 JAA93792.1 KA649718 AFP64347.1 GEDC01022718 JAS14580.1 KA649578 KA649719 AFP64348.1 J04567 X14179 X67650 X61223 X61224 AE014134 AY058492 X80204 M13786 J03927 CM002910 KMY88404.1 AGB92649.1 CM000157 KRJ98068.1 KA646634 AFP61263.1 AGB92647.1 EDW88874.1 CH902620 KPU73708.1 CM000361 EDX03874.1 KMY88403.1 BT044153 ACH92218.1 AGB92648.1 EDV31931.1 OUUW01000010 SPP86067.1 CH954177 KQS70198.1 CH916368 EDW03020.1 EDV57920.1 U59682 CH379058 AY754559 L41249 U47885 AB019506 AB019507 D50091 BAA20577.1
ATU31389.1 KQ459606 KPI91302.1 KQ461133 KPJ08866.1 AGBW02007748 OWR54615.1 NWSH01002106 PCG69167.1 PCG69168.1 KF170736 AGW15345.1 ODYU01005592 SOQ46581.1 GDAI01001486 JAI16117.1 KQ971312 KYB29132.1 KYB29133.1 EEZ98249.2 GALX01004003 JAB64463.1 GAMD01002723 JAA98867.1 GGFL01004735 MBW68913.1 GGFM01002002 MBW22753.1 GGFK01002416 MBW35737.1 GGFK01002415 MBW35736.1 ADMH02000065 ETN67939.1 AXCN02000594 CH477221 EAT47333.1 GGFM01001967 MBW22718.1 APGK01048501 KB741112 ENN73826.1 GFDF01001832 JAV12252.1 EAT47334.1 ATLV01017808 KE525192 KFB42472.1 GEZM01044288 JAV78327.1 CVRI01000073 CRL07613.1 AJWK01008357 AJWK01008358 BT128609 AEE63566.1 KQ435762 KOX75460.1 EAT47332.1 JXUM01011468 JXUM01011469 JXUM01011470 JXUM01011471 KQ560331 KXJ83002.1 GANO01001833 JAB58038.1 GL445725 EFN89159.1 GAPW01002322 JAC11276.1 KB632184 ERL89623.1 NNAY01000716 OXU26833.1 AAAB01008807 EDO64530.1 APCN01000215 EAA03917.3 GFDL01006826 JAV28219.1 DS231815 EDS35205.1 ADTU01003700 AXCM01000512 JRES01000501 KNC30725.1 LJIG01002821 KRT84119.1 AY438978 AAR13229.1 GL441497 EFN64733.1 GAMC01019481 JAB87074.1 CH963857 EDW76352.1 GFDG01000838 JAV17961.1 GFDG01003888 JAV14911.1 KK107019 QOIP01000007 EZA62634.1 RLU20422.1 GAMC01019478 JAB87077.1 GALA01001060 JAA93792.1 KA649718 AFP64347.1 GEDC01022718 JAS14580.1 KA649578 KA649719 AFP64348.1 J04567 X14179 X67650 X61223 X61224 AE014134 AY058492 X80204 M13786 J03927 CM002910 KMY88404.1 AGB92649.1 CM000157 KRJ98068.1 KA646634 AFP61263.1 AGB92647.1 EDW88874.1 CH902620 KPU73708.1 CM000361 EDX03874.1 KMY88403.1 BT044153 ACH92218.1 AGB92648.1 EDV31931.1 OUUW01000010 SPP86067.1 CH954177 KQS70198.1 CH916368 EDW03020.1 EDV57920.1 U59682 CH379058 AY754559 L41249 U47885 AB019506 AB019507 D50091 BAA20577.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000076408
+ More
UP000007266 UP000000673 UP000069272 UP000075886 UP000075880 UP000075900 UP000008820 UP000075885 UP000019118 UP000030765 UP000183832 UP000092461 UP000053105 UP000069940 UP000249989 UP000008237 UP000075920 UP000030742 UP000002358 UP000215335 UP000007062 UP000075840 UP000075903 UP000075882 UP000002320 UP000076407 UP000005205 UP000075883 UP000037069 UP000075884 UP000095300 UP000000311 UP000007798 UP000053097 UP000279307 UP000095301 UP000000803 UP000002282 UP000007801 UP000000304 UP000268350 UP000008711 UP000001070 UP000001819 UP000192221
UP000007266 UP000000673 UP000069272 UP000075886 UP000075880 UP000075900 UP000008820 UP000075885 UP000019118 UP000030765 UP000183832 UP000092461 UP000053105 UP000069940 UP000249989 UP000008237 UP000075920 UP000030742 UP000002358 UP000215335 UP000007062 UP000075840 UP000075903 UP000075882 UP000002320 UP000076407 UP000005205 UP000075883 UP000037069 UP000075884 UP000095300 UP000000311 UP000007798 UP000053097 UP000279307 UP000095301 UP000000803 UP000002282 UP000007801 UP000000304 UP000268350 UP000008711 UP000001070 UP000001819 UP000192221
Pfam
Interpro
IPR036291
NAD(P)-bd_dom_sf
+ More
IPR006168 G3P_DH_NAD-dep
IPR008927 6-PGluconate_DH-like_C_sf
IPR011128 G3P_DH_NAD-dep_N
IPR013328 6PGD_dom2
IPR017751 G3P_DH_NAD-dep_euk
IPR006109 G3P_DH_NAD-dep_C
IPR018593 tRNA-endonuc_su_Sen15
IPR022164 Kinesin-like
IPR000253 FHA_dom
IPR036859 CAP-Gly_dom_sf
IPR036167 tRNA_intron_Endo_cat-like_sf
IPR008984 SMAD_FHA_dom_sf
IPR011856 tRNA_endonuc-like_dom_sf
IPR032923 KIF13A
IPR022140 Kinesin-like_KIF1-typ
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR032405 Kinesin_assoc
IPR036961 Kinesin_motor_dom_sf
IPR000938 CAP-Gly_domain
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR013026 TPR-contain_dom
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR021183 NatA_aux_su
IPR007757 MT-A70-like
IPR029063 SAM-dependent_MTases
IPR002052 DNA_methylase_N6_adenine_CS
IPR006168 G3P_DH_NAD-dep
IPR008927 6-PGluconate_DH-like_C_sf
IPR011128 G3P_DH_NAD-dep_N
IPR013328 6PGD_dom2
IPR017751 G3P_DH_NAD-dep_euk
IPR006109 G3P_DH_NAD-dep_C
IPR018593 tRNA-endonuc_su_Sen15
IPR022164 Kinesin-like
IPR000253 FHA_dom
IPR036859 CAP-Gly_dom_sf
IPR036167 tRNA_intron_Endo_cat-like_sf
IPR008984 SMAD_FHA_dom_sf
IPR011856 tRNA_endonuc-like_dom_sf
IPR032923 KIF13A
IPR022140 Kinesin-like_KIF1-typ
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR032405 Kinesin_assoc
IPR036961 Kinesin_motor_dom_sf
IPR000938 CAP-Gly_domain
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR013026 TPR-contain_dom
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR021183 NatA_aux_su
IPR007757 MT-A70-like
IPR029063 SAM-dependent_MTases
IPR002052 DNA_methylase_N6_adenine_CS
SUPFAM
CDD
ProteinModelPortal
Q68AX4
Q68AX3
H9JSP9
A0A2D3E1P0
A0A194PDH5
A0A0N1PI50
+ More
A0A212FLJ1 A0A2A4JAM7 A0A2A4JBU8 U3MGC9 A0A2H1W0H9 A0A0K8TP17 A0A182XZ58 A0A139WMI0 A0A139WMM5 D6W8W3 V5GW65 T1E8J2 A0A2M4CUC1 A0A2M3Z2N7 A0A2M4A4L1 A0A2M4A4J8 W5JX93 A0A182FI57 A0A182QY36 A0A182IJN4 A0A182RHS9 Q17KS5 A0A1S4EZ75 A0A182P3N4 A0A2M3Z2P5 N6TZN6 A0A1L8E105 Q17KS3 A0A084VWX8 A0A1Y1M526 A0A1J1J5C4 A0A1B0CDX8 J3JZC8 A0A0M9A1K4 Q17KS4 A0A182GGV8 U5EKF5 E2B565 A0A182WIL2 A0A023ERI3 U4UBB4 K7IYD0 A0A232F8R0 A7UR88 A0A182I3L1 A0A182V4T7 Q7PTH2 A0A1Q3FKV5 A0A182KRQ9 B0VZW3 A0A182X3F7 A0A158NXX9 A0A182MJ04 A0A0L0CEP4 A0A0T6BB32 A0A1Y9H2M9 A0A1I8Q1S4 A0A1I8Q1T4 Q6T312 A0A1I8Q1V2 E2AP99 W8BB57 B4MV07 A0A1L8EGZ8 A0A1L8E884 A0A026X5E1 W8AMA6 T1E2R4 A0A1I8Q1V3 T1PP02 A0A1B6CMA5 T1PLW5 P13706-2 A0A0J9QXF0 M9PC43 A0A0R1DLB1 T1PD78 P13706 P13706-4 P13706-3 M9PET0 B4NZU0 A0A0N8NZ96 B4Q441 B5RIM9 B3MMP0 A0A3B0JVE2 A0A0Q5W9H8 B4JBT5 B3N590 Q27928-3 O97463-2 A0A1W4UNC2 Q9TYF7
A0A212FLJ1 A0A2A4JAM7 A0A2A4JBU8 U3MGC9 A0A2H1W0H9 A0A0K8TP17 A0A182XZ58 A0A139WMI0 A0A139WMM5 D6W8W3 V5GW65 T1E8J2 A0A2M4CUC1 A0A2M3Z2N7 A0A2M4A4L1 A0A2M4A4J8 W5JX93 A0A182FI57 A0A182QY36 A0A182IJN4 A0A182RHS9 Q17KS5 A0A1S4EZ75 A0A182P3N4 A0A2M3Z2P5 N6TZN6 A0A1L8E105 Q17KS3 A0A084VWX8 A0A1Y1M526 A0A1J1J5C4 A0A1B0CDX8 J3JZC8 A0A0M9A1K4 Q17KS4 A0A182GGV8 U5EKF5 E2B565 A0A182WIL2 A0A023ERI3 U4UBB4 K7IYD0 A0A232F8R0 A7UR88 A0A182I3L1 A0A182V4T7 Q7PTH2 A0A1Q3FKV5 A0A182KRQ9 B0VZW3 A0A182X3F7 A0A158NXX9 A0A182MJ04 A0A0L0CEP4 A0A0T6BB32 A0A1Y9H2M9 A0A1I8Q1S4 A0A1I8Q1T4 Q6T312 A0A1I8Q1V2 E2AP99 W8BB57 B4MV07 A0A1L8EGZ8 A0A1L8E884 A0A026X5E1 W8AMA6 T1E2R4 A0A1I8Q1V3 T1PP02 A0A1B6CMA5 T1PLW5 P13706-2 A0A0J9QXF0 M9PC43 A0A0R1DLB1 T1PD78 P13706 P13706-4 P13706-3 M9PET0 B4NZU0 A0A0N8NZ96 B4Q441 B5RIM9 B3MMP0 A0A3B0JVE2 A0A0Q5W9H8 B4JBT5 B3N590 Q27928-3 O97463-2 A0A1W4UNC2 Q9TYF7
PDB
1X0X
E-value=4.49075e-128,
Score=1172
Ontologies
GO
GO:0042803
GO:0004367
GO:0005975
GO:0009331
GO:0051287
GO:0046168
GO:0008017
GO:0003777
GO:0005524
GO:0003676
GO:0000213
GO:0006388
GO:0007018
GO:0005829
GO:0006072
GO:0006116
GO:0016021
GO:0008168
GO:0006641
GO:0055093
GO:0007629
GO:0055114
GO:0006067
GO:0030018
GO:0006650
GO:0031430
GO:0016616
GO:0016491
GO:0003824
PANTHER
Topology
Subcellular location
Cytoplasm
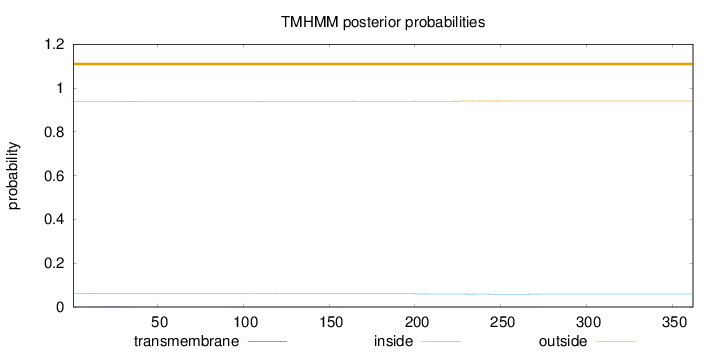
Length:
362
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10277
Exp number, first 60 AAs:
0.04483
Total prob of N-in:
0.06272
outside
1 - 362
Population Genetic Test Statistics
Pi
232.803312
Theta
193.773542
Tajima's D
0.587675
CLR
0.070148
CSRT
0.540522973851307
Interpretation
Uncertain
