Gene
KWMTBOMO05053
Pre Gene Modal
BGIBMGA012539
Annotation
PREDICTED:_opsin?_ultraviolet-sensitive_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.888
Sequence
CDS
ATGGATACAGTGCCAAAAGGTATAAACAATTTAACGTATTTGAAAAGAGATGATCATGCTCTATTCAGTTCTAAATATGACGATGAAACTCTTACGAAAGGTGACATAAAATTGTGGAAAAGACGTCGCAGAATGTCTGAAATTTTGGACAGAAGATCAAATGTAAGCGGTGAGGTCAAAGAAACTAGACTACCAATAAAGTATTCCAAGTACGAAGTGAATGAATTGGTTTCATCGGAAGCTAACAACGAGGTATTAATAAAAAAATTCAAAGAACTTTGGCCAGTAAATTTATGGCGTCGCTTTGGCCTGTTTACCGATGATTATCTACACTTGGTGAATCCATACTGGCTGAGCTTCGCCCCGCCACATCCTTTACTGCATTATGGCCTCGGTGCATTCTACATCATGATGACCACTGTTGGGTCCACTGGCAATGCTTTGGTGATGTTCATGTATTTTAGATGCCGTTCCCTGAGAACGCCGGGAAATATTCTAGTTGCAAATCTAGCATTGAGTGATTTTATGATGCTTGCGAAGACACCTATTTTTATATTTAATTCGTTTAACCTCGGACCAGCTTTAGGCAAAACTGGTTGCGTTATTTATGGATTTGTTGGTGGGTTAACAGGAACTACGTCAATAGCAATCCTAACCGCAATATCCTTAGATCGGTACTGGGCGGTCGTGCGGCCATTGGAACCACTACGAGCTCTGACTGCCATCCGTGCTCGACTCATGGCAATAGCTGCATGGATTTACGCTACATTATTTGCCATAATTCCAGCCTTGGATATAGGTTACGGTCGCTACGTGCCCGAAGGATTCCTGACAAGCTGTAGTTTTGATTATCTAACTGAAGAACTTCCGCCTCGCTATTTCATTTTTGTATTTTTTTGCGCAGCCTGGCTCGCGCCTTTCTGCACCATATCGTACTGTTATATAAGCATATTTCGGGTTGTCGTATGTAGTCGAAATATTACTACAAAAAATCAAGAACAACGGCTCTCCACGAGACATGTAAAGGAGCGAACAAAACGTAAAGCTGAAATCAAACTTGCCTTTTTGGTTATAGTTGTAATTGCATTGTTTTTTATTTCATGGACACCGTATGCAATTGTTGCACTTTTAGGAATATTTGGAAAAAAGGATCTTATCAAACCGCTAACATCCATGATACCTGCATTATTTTGTAAAACAGCCGCTTGTATTAATCCTTTCATTTATATCATTACTCATCCTAAATTTCGAAAGGAATTCAAGAGAATGTTGTCCAGAGATAAAACAGAACGAAGTGGAACAATAAAAACTATTGGATATTATACTGAAGCGAGTAAATTCCATCGTCCTAGTAAGGACTTCAGTGACACTGAAGTTGAAATAGTTGAAATGAAAGATATACCGTACCGAAATGAAGACACAACTAACACAGTTGAAGAAAAAATTAATACGGTGTCTGCTAAATTAGCTAAACGAGAAGGATCGCAAAAATCGCAAAGTAGTATCAGCATGAAAAGTTTCGAAGAAAACGTTGTGAGTCCCCCTTCGTGGTATTGTAAACCGAAATTCGCCAAAAAAAAGAGTTTCCATCGTCGTTCTACACATAGTATGGTTTCTCAAAATACCGATCCAACTGTCTAA
Protein
MDTVPKGINNLTYLKRDDHALFSSKYDDETLTKGDIKLWKRRRRMSEILDRRSNVSGEVKETRLPIKYSKYEVNELVSSEANNEVLIKKFKELWPVNLWRRFGLFTDDYLHLVNPYWLSFAPPHPLLHYGLGAFYIMMTTVGSTGNALVMFMYFRCRSLRTPGNILVANLALSDFMMLAKTPIFIFNSFNLGPALGKTGCVIYGFVGGLTGTTSIAILTAISLDRYWAVVRPLEPLRALTAIRARLMAIAAWIYATLFAIIPALDIGYGRYVPEGFLTSCSFDYLTEELPPRYFIFVFFCAAWLAPFCTISYCYISIFRVVVCSRNITTKNQEQRLSTRHVKERTKRKAEIKLAFLVIVVIALFFISWTPYAIVALLGIFGKKDLIKPLTSMIPALFCKTAACINPFIYIITHPKFRKEFKRMLSRDKTERSGTIKTIGYYTEASKFHRPSKDFSDTEVEIVEMKDIPYRNEDTTNTVEEKINTVSAKLAKREGSQKSQSSISMKSFEENVVSPPSWYCKPKFAKKKSFHRRSTHSMVSQNTDPTV
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family. Opsin subfamily.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 1 family.
Uniprot
EMBL
BABH01032141
RSAL01000007
RVE54062.1
NWSH01002106
PCG69166.1
ODYU01012934
+ More
SOQ59476.1 KQ459606 KPI91300.1 JTDY01006086 KOB66307.1 GGMR01014084 MBY26703.1 GGMS01005880 MBY75083.1 MF434481 AWM72040.1 ABLF02040785 ABLF02040802 ABLF02040811 ABLF02040817 ABLF02040819 ABLF02040820 ABLF02040832 MF434482 AWM72041.1 LC009260 BAQ54908.1 LC009237 BAQ54884.1 LC009099 BAQ54741.1 LC009123 BAQ54766.1 LC009059 BAQ54697.1 BK008815 DAA64804.1 CH916366 EDV96260.1 CVRI01000067 CRL06422.1
SOQ59476.1 KQ459606 KPI91300.1 JTDY01006086 KOB66307.1 GGMR01014084 MBY26703.1 GGMS01005880 MBY75083.1 MF434481 AWM72040.1 ABLF02040785 ABLF02040802 ABLF02040811 ABLF02040817 ABLF02040819 ABLF02040820 ABLF02040832 MF434482 AWM72041.1 LC009260 BAQ54908.1 LC009237 BAQ54884.1 LC009099 BAQ54741.1 LC009123 BAQ54766.1 LC009059 BAQ54697.1 BK008815 DAA64804.1 CH916366 EDV96260.1 CVRI01000067 CRL06422.1
Proteomes
PRIDE
Interpro
IPR001760
Opsin
+ More
IPR017452 GPCR_Rhodpsn_7TM
IPR027430 Retinal_BS
IPR000276 GPCR_Rhodpsn
IPR000856 Opsin_RH3/RH4
IPR041711 Met-tRNA-FMT_N
IPR036477 Formyl_transf_N_sf
IPR002376 Formyl_transf_N
IPR037022 Formyl_trans_C_sf
IPR011034 Formyl_transferase-like_C_sf
IPR005794 Fmt
IPR005793 Formyl_trans_C
IPR017452 GPCR_Rhodpsn_7TM
IPR027430 Retinal_BS
IPR000276 GPCR_Rhodpsn
IPR000856 Opsin_RH3/RH4
IPR041711 Met-tRNA-FMT_N
IPR036477 Formyl_transf_N_sf
IPR002376 Formyl_transf_N
IPR037022 Formyl_trans_C_sf
IPR011034 Formyl_transferase-like_C_sf
IPR005794 Fmt
IPR005793 Formyl_trans_C
Gene 3D
ProteinModelPortal
PDB
3AYN
E-value=6.54343e-47,
Score=474
Ontologies
GO
Topology
Subcellular location
Membrane
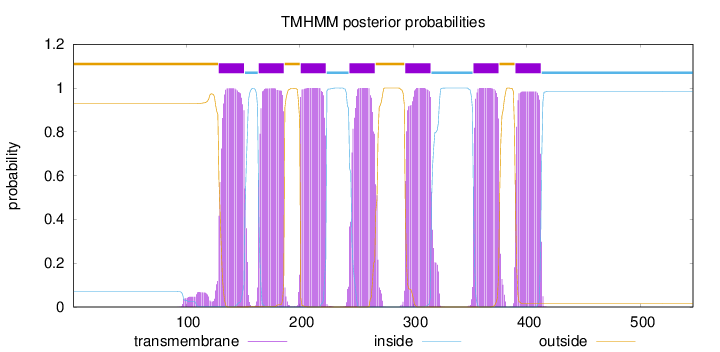
Length:
546
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
160.67882
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07006
outside
1 - 128
TMhelix
129 - 151
inside
152 - 163
TMhelix
164 - 186
outside
187 - 200
TMhelix
201 - 223
inside
224 - 243
TMhelix
244 - 266
outside
267 - 292
TMhelix
293 - 315
inside
316 - 352
TMhelix
353 - 375
outside
376 - 389
TMhelix
390 - 412
inside
413 - 546
Population Genetic Test Statistics
Pi
258.779833
Theta
199.826356
Tajima's D
1.019334
CLR
0
CSRT
0.661816909154542
Interpretation
Uncertain
