Gene
KWMTBOMO05047
Pre Gene Modal
BGIBMGA012565
Annotation
PREDICTED:_decaprenyl-diphosphate_synthase_subunit_1_isoform_X1_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.933
Sequence
CDS
ATGGCGTGTTTAAGTTCGAGTAGGCTCTGTGCTCGTGTTCAGGATAAGTTAATGTGTCAATTTGGACATAAACTTCGAAAACAAACTGTATTTGTGAAACTCAAACCTCCTGGACCATCTCCGTTGCGGACATTTGGAAAGACATGTCAGACGGTCACGACATGCGGTCCGACATGTGGAGCCGGCAGTTTACCGGTCCCCGGCGGCTTAGTGCGCCGCTTCGCCTCCAATTTACAGAGCACAAGCACCGGCTCTCTGCCGGATTACGGCACACAGATCATTGATCCATATCGCTTATTAGAAGATGACCTTAATGGAATATATGAGGATATTAGATCGGAACTAGAAAGAAACACAAACCAATCAGAATTGAACACCATAGCCACATACTACTTCGACGGACAAGGCAAGGCACTCAGACCGATGGTTGCGATTTTAATGGCCAGGGCTGTCAACTACCACGTTTACGGAGAGAACAGTGCACTGCTACCGTCGCAGCGACAAGTGGCGATGATCAGTGAGATGATCCACTCGGCGTCTCTGATCCACGATGATGTCATCGACCAAAGCGACTTCCGTCGCGGGAAACCCTCCGTCAACGTGCTCTGGAATCACAAAAAGGTGGCGATGGCCGGCGACTTCATCCTCGCTGTGGCGTCGATGATGATAGCTCGCTTGCGCAGCGATGATGTCACACTCGTGCTCAGTCAAGTGGTAACAGATTTGGTGCAGGGCGAATTCATGCAACTTGGCAGCAAAGAAACTGAAAACGAGCGGTTCGCTCACTACCTCACAAAGACGTACAGGAAGACGGCTTCATTAATTGCTAATTCCGTTAAGGCGGTCGCCTTACTGAGCGGCGCCGACGAGAGCACGTGCGAGTTGGCGTTCCAATACGGACGGAACTTGGGGCTCGCGTTTCAGCTCGTCGACGACTTGTTGGACTTCGTGTCTTCAGCACAGGCTATGGGCAAGCCCACCGCGGCCGACCTCAAGCTCGGTCTTGCCACCGCGCCTGTTCTATTCGCCTGCGAAAAGTATCCGGAGCTGAATCCGATGATAATGAGGCGGTTCCAAGAGTCCGGCGACGTGGAGAAGGCGTTCGAATTGGTCCACAAGTCTCGCGGCCTCGAGCAGACGCGTTTCCTAGCGCGCAAGCACGGCGCCGAGGCCGCGAGACTGGCGGCCGACCTGGCTGACTCGCCCTACCAGAAGGGACTAGTGCTCACCACCGACCTCGTCCTCAACAGGATCAAATAG
Protein
MACLSSSRLCARVQDKLMCQFGHKLRKQTVFVKLKPPGPSPLRTFGKTCQTVTTCGPTCGAGSLPVPGGLVRRFASNLQSTSTGSLPDYGTQIIDPYRLLEDDLNGIYEDIRSELERNTNQSELNTIATYYFDGQGKALRPMVAILMARAVNYHVYGENSALLPSQRQVAMISEMIHSASLIHDDVIDQSDFRRGKPSVNVLWNHKKVAMAGDFILAVASMMIARLRSDDVTLVLSQVVTDLVQGEFMQLGSKETENERFAHYLTKTYRKTASLIANSVKAVALLSGADESTCELAFQYGRNLGLAFQLVDDLLDFVSSAQAMGKPTAADLKLGLATAPVLFACEKYPELNPMIMRRFQESGDVEKAFELVHKSRGLEQTRFLARKHGAEAARLAADLADSPYQKGLVLTTDLVLNRIK
Summary
Similarity
Belongs to the FPP/GGPP synthase family.
Uniprot
A0A212F9C8
A0A0N1PGS1
G9D512
A0A2A4JK80
A0A1B6KSJ0
A0A1B6GMQ4
+ More
A0A1B6DIQ4 A0A1Y1MX09 A0A0C9Q793 A0A154P407 K7JBK8 D6WAF3 A0A0L7QLJ7 A0A232ET71 A0A067QX52 A0A224XQ55 A0A023F505 A0A1L8DZU4 J9K4T0 A0A1L8DZQ2 V5UUH0 A0A0T6AYV0 A0A2H8TKV8 A0A2C9H5R7 M9VU49 Q5TV64 A0A2C9GPJ6 A0A182JJT3 A0A2M4A8N7 A0A2M3Z5T0 A0A2M4A8N0 A0A2M3Z5P8 A0A084VNV0 A0A2M4BNT9 A0A026VYX6 A0A2M4CUI7 A0A2M4CUG1 A0A2M4A527 A0A0L0CEU7 A0A2M3Z370 A0A0A1WFE6 A0A2M4BKU5 A0A0M5J5L1 B3P7R1 A0A195DY43 B4GNS0 A0A2M4BJ41 A0A1W4UJU4 Q29BC6 B4PNB8 W8BDU2 A0A3B0K6B0 A0A034VA95 A0A0K8UDS6 Q9V9Z3 A0A195B7S7 F4WZX1 B4LW00 B3M080 A0A1Q3FU52 B4NH17 A0A1Q3FUM2 A0A1Q3FU57 A0A1Q3FUI1 A0A1A9VVE4 A0A1Q3FU35 A0A1B0CK66 A0A1Q3FU59 A0A1Q3FU96 A0A1Q3FUL0 B4JI23 B4K4J3 A0A1B0G312 T1P8F4 A0A1A9XTG1 A0A1A9ZJJ3 E2AMF4 A0A3L8DMT1 A0A182Q2S2 A0A1J1HPS1 A0A151XAF4 A0A1I8PDC6 A0A0K8VLI7 B4I007 B4QSK9 A0A0K8UWD0 A0A0K8SQK6 E2C655 A0A182P568 A0A182VZ71 A0A182R669 A0A182JSE6 A0A195FEF4 A0A151ICL0 A0A1B6JR16 A0A182MTW1 A0A182YAP7 A0A182X0B1 A0A0A9ZEP0 A0A182TH73
A0A1B6DIQ4 A0A1Y1MX09 A0A0C9Q793 A0A154P407 K7JBK8 D6WAF3 A0A0L7QLJ7 A0A232ET71 A0A067QX52 A0A224XQ55 A0A023F505 A0A1L8DZU4 J9K4T0 A0A1L8DZQ2 V5UUH0 A0A0T6AYV0 A0A2H8TKV8 A0A2C9H5R7 M9VU49 Q5TV64 A0A2C9GPJ6 A0A182JJT3 A0A2M4A8N7 A0A2M3Z5T0 A0A2M4A8N0 A0A2M3Z5P8 A0A084VNV0 A0A2M4BNT9 A0A026VYX6 A0A2M4CUI7 A0A2M4CUG1 A0A2M4A527 A0A0L0CEU7 A0A2M3Z370 A0A0A1WFE6 A0A2M4BKU5 A0A0M5J5L1 B3P7R1 A0A195DY43 B4GNS0 A0A2M4BJ41 A0A1W4UJU4 Q29BC6 B4PNB8 W8BDU2 A0A3B0K6B0 A0A034VA95 A0A0K8UDS6 Q9V9Z3 A0A195B7S7 F4WZX1 B4LW00 B3M080 A0A1Q3FU52 B4NH17 A0A1Q3FUM2 A0A1Q3FU57 A0A1Q3FUI1 A0A1A9VVE4 A0A1Q3FU35 A0A1B0CK66 A0A1Q3FU59 A0A1Q3FU96 A0A1Q3FUL0 B4JI23 B4K4J3 A0A1B0G312 T1P8F4 A0A1A9XTG1 A0A1A9ZJJ3 E2AMF4 A0A3L8DMT1 A0A182Q2S2 A0A1J1HPS1 A0A151XAF4 A0A1I8PDC6 A0A0K8VLI7 B4I007 B4QSK9 A0A0K8UWD0 A0A0K8SQK6 E2C655 A0A182P568 A0A182VZ71 A0A182R669 A0A182JSE6 A0A195FEF4 A0A151ICL0 A0A1B6JR16 A0A182MTW1 A0A182YAP7 A0A182X0B1 A0A0A9ZEP0 A0A182TH73
Pubmed
22118469
26354079
21967427
28004739
20075255
18362917
+ More
19820115 28648823 24845553 25474469 24246678 12364791 14747013 17210077 24438588 24508170 26108605 25830018 17994087 15632085 17550304 24495485 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 18057021 25315136 20798317 30249741 26823975 25244985 25401762
19820115 28648823 24845553 25474469 24246678 12364791 14747013 17210077 24438588 24508170 26108605 25830018 17994087 15632085 17550304 24495485 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 18057021 25315136 20798317 30249741 26823975 25244985 25401762
EMBL
AGBW02009609
OWR50330.1
KQ461133
KPJ08874.1
JF777415
AEU11760.1
+ More
NWSH01001295 PCG71810.1 GEBQ01025580 JAT14397.1 GECZ01006041 JAS63728.1 GEDC01011783 JAS25515.1 GEZM01018559 JAV90173.1 GBYB01010018 JAG79785.1 KQ434803 KZC05958.1 AAZX01008745 AAZX01020998 KQ971312 EEZ98009.2 KQ414915 KOC59495.1 NNAY01002346 OXU21496.1 KK852865 KDR14770.1 GFTR01006153 JAW10273.1 GBBI01002116 JAC16596.1 GFDF01002168 JAV11916.1 ABLF02029495 ABLF02029496 GFDF01002167 JAV11917.1 KF673861 AHB81557.1 LJIG01022495 KRT80273.1 GFXV01002972 MBW14777.1 KC431243 AGJ83979.1 AAAB01008846 EAL41155.4 APCN01001103 APCN01001104 GGFK01003836 MBW37157.1 GGFM01003104 MBW23855.1 GGFK01003804 MBW37125.1 GGFM01003096 MBW23847.1 ATLV01014925 ATLV01014926 ATLV01014927 ATLV01014928 ATLV01014929 KE524996 KFB39644.1 GGFJ01005574 MBW54715.1 KK107681 EZA48064.1 GGFL01004805 MBW68983.1 GGFL01004806 MBW68984.1 GGFK01002534 MBW35855.1 JRES01000501 KNC30742.1 GGFM01002216 MBW22967.1 GBXI01017154 JAC97137.1 GGFJ01004549 MBW53690.1 CP012526 ALC47961.1 CH954182 EDV52969.1 KQ980107 KYN17627.1 CH479186 EDW38803.1 GGFJ01003926 MBW53067.1 CM000070 EAL27072.2 CM000160 EDW99200.2 GAMC01011394 GAMC01011392 JAB95163.1 OUUW01000005 SPP81156.1 GAKP01020464 GAKP01020462 GAKP01020456 GAKP01020452 JAC38488.1 GDHF01032040 GDHF01027492 GDHF01024485 GDHF01017285 GDHF01015602 GDHF01007310 GDHF01004066 GDHF01002098 GDHF01002061 JAI20274.1 JAI24822.1 JAI27829.1 JAI35029.1 JAI36712.1 JAI45004.1 JAI48248.1 JAI50216.1 JAI50253.1 AE014297 BT044087 AAF57135.2 ACH92152.1 AHN57618.1 KQ976574 KYM80300.1 GL888480 EGI60261.1 CH940650 EDW66505.1 KRF82727.1 CH902617 EDV42037.1 GFDL01003916 JAV31129.1 CH964272 EDW84514.1 GFDL01003912 JAV31133.1 GFDL01003915 JAV31130.1 GFDL01003909 JAV31136.1 GFDL01003914 JAV31131.1 AJWK01015817 AJWK01015818 GFDL01003911 JAV31134.1 GFDL01003908 JAV31137.1 GFDL01003913 JAV31132.1 CH916369 EDV93943.1 CH933806 EDW13945.1 CCAG010021770 KA644919 AFP59548.1 GL440813 EFN65378.1 QOIP01000006 RLU21586.1 AXCN02001244 CVRI01000015 CRK89963.1 KQ982339 KYQ57343.1 GDHF01031272 GDHF01012869 JAI21042.1 JAI39445.1 CH480819 EDW53614.1 CM000364 EDX15106.1 GDHF01021428 JAI30886.1 GBRD01010714 GDHC01009406 JAG55110.1 JAQ09223.1 GL452926 EFN76575.1 KQ981625 KYN39060.1 KQ978053 KYM97779.1 GECU01006363 JAT01344.1 AXCM01012380 AXCM01012381 GBHO01001771 JAG41833.1
NWSH01001295 PCG71810.1 GEBQ01025580 JAT14397.1 GECZ01006041 JAS63728.1 GEDC01011783 JAS25515.1 GEZM01018559 JAV90173.1 GBYB01010018 JAG79785.1 KQ434803 KZC05958.1 AAZX01008745 AAZX01020998 KQ971312 EEZ98009.2 KQ414915 KOC59495.1 NNAY01002346 OXU21496.1 KK852865 KDR14770.1 GFTR01006153 JAW10273.1 GBBI01002116 JAC16596.1 GFDF01002168 JAV11916.1 ABLF02029495 ABLF02029496 GFDF01002167 JAV11917.1 KF673861 AHB81557.1 LJIG01022495 KRT80273.1 GFXV01002972 MBW14777.1 KC431243 AGJ83979.1 AAAB01008846 EAL41155.4 APCN01001103 APCN01001104 GGFK01003836 MBW37157.1 GGFM01003104 MBW23855.1 GGFK01003804 MBW37125.1 GGFM01003096 MBW23847.1 ATLV01014925 ATLV01014926 ATLV01014927 ATLV01014928 ATLV01014929 KE524996 KFB39644.1 GGFJ01005574 MBW54715.1 KK107681 EZA48064.1 GGFL01004805 MBW68983.1 GGFL01004806 MBW68984.1 GGFK01002534 MBW35855.1 JRES01000501 KNC30742.1 GGFM01002216 MBW22967.1 GBXI01017154 JAC97137.1 GGFJ01004549 MBW53690.1 CP012526 ALC47961.1 CH954182 EDV52969.1 KQ980107 KYN17627.1 CH479186 EDW38803.1 GGFJ01003926 MBW53067.1 CM000070 EAL27072.2 CM000160 EDW99200.2 GAMC01011394 GAMC01011392 JAB95163.1 OUUW01000005 SPP81156.1 GAKP01020464 GAKP01020462 GAKP01020456 GAKP01020452 JAC38488.1 GDHF01032040 GDHF01027492 GDHF01024485 GDHF01017285 GDHF01015602 GDHF01007310 GDHF01004066 GDHF01002098 GDHF01002061 JAI20274.1 JAI24822.1 JAI27829.1 JAI35029.1 JAI36712.1 JAI45004.1 JAI48248.1 JAI50216.1 JAI50253.1 AE014297 BT044087 AAF57135.2 ACH92152.1 AHN57618.1 KQ976574 KYM80300.1 GL888480 EGI60261.1 CH940650 EDW66505.1 KRF82727.1 CH902617 EDV42037.1 GFDL01003916 JAV31129.1 CH964272 EDW84514.1 GFDL01003912 JAV31133.1 GFDL01003915 JAV31130.1 GFDL01003909 JAV31136.1 GFDL01003914 JAV31131.1 AJWK01015817 AJWK01015818 GFDL01003911 JAV31134.1 GFDL01003908 JAV31137.1 GFDL01003913 JAV31132.1 CH916369 EDV93943.1 CH933806 EDW13945.1 CCAG010021770 KA644919 AFP59548.1 GL440813 EFN65378.1 QOIP01000006 RLU21586.1 AXCN02001244 CVRI01000015 CRK89963.1 KQ982339 KYQ57343.1 GDHF01031272 GDHF01012869 JAI21042.1 JAI39445.1 CH480819 EDW53614.1 CM000364 EDX15106.1 GDHF01021428 JAI30886.1 GBRD01010714 GDHC01009406 JAG55110.1 JAQ09223.1 GL452926 EFN76575.1 KQ981625 KYN39060.1 KQ978053 KYM97779.1 GECU01006363 JAT01344.1 AXCM01012380 AXCM01012381 GBHO01001771 JAG41833.1
Proteomes
UP000007151
UP000053240
UP000218220
UP000076502
UP000002358
UP000007266
+ More
UP000053825 UP000215335 UP000027135 UP000007819 UP000075903 UP000007062 UP000075840 UP000075880 UP000030765 UP000053097 UP000037069 UP000092553 UP000008711 UP000078492 UP000008744 UP000192221 UP000001819 UP000002282 UP000268350 UP000000803 UP000078540 UP000007755 UP000008792 UP000007801 UP000007798 UP000078200 UP000092461 UP000001070 UP000009192 UP000092444 UP000095301 UP000092443 UP000092445 UP000000311 UP000279307 UP000075886 UP000183832 UP000075809 UP000095300 UP000001292 UP000000304 UP000008237 UP000075885 UP000075920 UP000075900 UP000075881 UP000078541 UP000078542 UP000075883 UP000076408 UP000076407 UP000075902
UP000053825 UP000215335 UP000027135 UP000007819 UP000075903 UP000007062 UP000075840 UP000075880 UP000030765 UP000053097 UP000037069 UP000092553 UP000008711 UP000078492 UP000008744 UP000192221 UP000001819 UP000002282 UP000268350 UP000000803 UP000078540 UP000007755 UP000008792 UP000007801 UP000007798 UP000078200 UP000092461 UP000001070 UP000009192 UP000092444 UP000095301 UP000092443 UP000092445 UP000000311 UP000279307 UP000075886 UP000183832 UP000075809 UP000095300 UP000001292 UP000000304 UP000008237 UP000075885 UP000075920 UP000075900 UP000075881 UP000078541 UP000078542 UP000075883 UP000076408 UP000076407 UP000075902
PRIDE
Interpro
SUPFAM
SSF48576
SSF48576
Gene 3D
ProteinModelPortal
A0A212F9C8
A0A0N1PGS1
G9D512
A0A2A4JK80
A0A1B6KSJ0
A0A1B6GMQ4
+ More
A0A1B6DIQ4 A0A1Y1MX09 A0A0C9Q793 A0A154P407 K7JBK8 D6WAF3 A0A0L7QLJ7 A0A232ET71 A0A067QX52 A0A224XQ55 A0A023F505 A0A1L8DZU4 J9K4T0 A0A1L8DZQ2 V5UUH0 A0A0T6AYV0 A0A2H8TKV8 A0A2C9H5R7 M9VU49 Q5TV64 A0A2C9GPJ6 A0A182JJT3 A0A2M4A8N7 A0A2M3Z5T0 A0A2M4A8N0 A0A2M3Z5P8 A0A084VNV0 A0A2M4BNT9 A0A026VYX6 A0A2M4CUI7 A0A2M4CUG1 A0A2M4A527 A0A0L0CEU7 A0A2M3Z370 A0A0A1WFE6 A0A2M4BKU5 A0A0M5J5L1 B3P7R1 A0A195DY43 B4GNS0 A0A2M4BJ41 A0A1W4UJU4 Q29BC6 B4PNB8 W8BDU2 A0A3B0K6B0 A0A034VA95 A0A0K8UDS6 Q9V9Z3 A0A195B7S7 F4WZX1 B4LW00 B3M080 A0A1Q3FU52 B4NH17 A0A1Q3FUM2 A0A1Q3FU57 A0A1Q3FUI1 A0A1A9VVE4 A0A1Q3FU35 A0A1B0CK66 A0A1Q3FU59 A0A1Q3FU96 A0A1Q3FUL0 B4JI23 B4K4J3 A0A1B0G312 T1P8F4 A0A1A9XTG1 A0A1A9ZJJ3 E2AMF4 A0A3L8DMT1 A0A182Q2S2 A0A1J1HPS1 A0A151XAF4 A0A1I8PDC6 A0A0K8VLI7 B4I007 B4QSK9 A0A0K8UWD0 A0A0K8SQK6 E2C655 A0A182P568 A0A182VZ71 A0A182R669 A0A182JSE6 A0A195FEF4 A0A151ICL0 A0A1B6JR16 A0A182MTW1 A0A182YAP7 A0A182X0B1 A0A0A9ZEP0 A0A182TH73
A0A1B6DIQ4 A0A1Y1MX09 A0A0C9Q793 A0A154P407 K7JBK8 D6WAF3 A0A0L7QLJ7 A0A232ET71 A0A067QX52 A0A224XQ55 A0A023F505 A0A1L8DZU4 J9K4T0 A0A1L8DZQ2 V5UUH0 A0A0T6AYV0 A0A2H8TKV8 A0A2C9H5R7 M9VU49 Q5TV64 A0A2C9GPJ6 A0A182JJT3 A0A2M4A8N7 A0A2M3Z5T0 A0A2M4A8N0 A0A2M3Z5P8 A0A084VNV0 A0A2M4BNT9 A0A026VYX6 A0A2M4CUI7 A0A2M4CUG1 A0A2M4A527 A0A0L0CEU7 A0A2M3Z370 A0A0A1WFE6 A0A2M4BKU5 A0A0M5J5L1 B3P7R1 A0A195DY43 B4GNS0 A0A2M4BJ41 A0A1W4UJU4 Q29BC6 B4PNB8 W8BDU2 A0A3B0K6B0 A0A034VA95 A0A0K8UDS6 Q9V9Z3 A0A195B7S7 F4WZX1 B4LW00 B3M080 A0A1Q3FU52 B4NH17 A0A1Q3FUM2 A0A1Q3FU57 A0A1Q3FUI1 A0A1A9VVE4 A0A1Q3FU35 A0A1B0CK66 A0A1Q3FU59 A0A1Q3FU96 A0A1Q3FUL0 B4JI23 B4K4J3 A0A1B0G312 T1P8F4 A0A1A9XTG1 A0A1A9ZJJ3 E2AMF4 A0A3L8DMT1 A0A182Q2S2 A0A1J1HPS1 A0A151XAF4 A0A1I8PDC6 A0A0K8VLI7 B4I007 B4QSK9 A0A0K8UWD0 A0A0K8SQK6 E2C655 A0A182P568 A0A182VZ71 A0A182R669 A0A182JSE6 A0A195FEF4 A0A151ICL0 A0A1B6JR16 A0A182MTW1 A0A182YAP7 A0A182X0B1 A0A0A9ZEP0 A0A182TH73
PDB
3APZ
E-value=6.96884e-57,
Score=559
Ontologies
GO
PANTHER
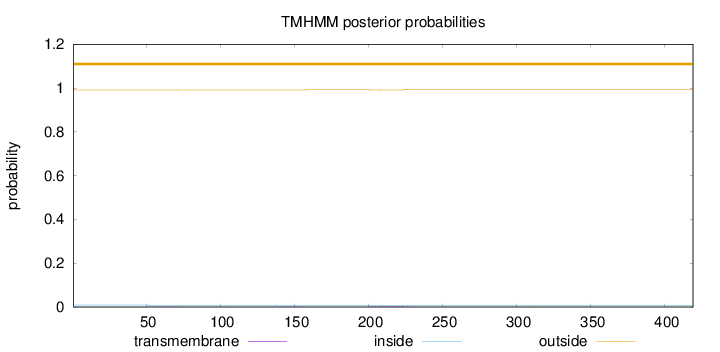
Topology
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06478
Exp number, first 60 AAs:
0.00234
Total prob of N-in:
0.00863
outside
1 - 419
Population Genetic Test Statistics
Pi
230.708339
Theta
186.462184
Tajima's D
0.466301
CLR
1.285922
CSRT
0.505224738763062
Interpretation
Uncertain
