Gene
KWMTBOMO05043
Annotation
PREDICTED:_carbonic_anhydrase_7-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.261
Sequence
CDS
ATGCGGTCTGTATGTTTAGTACTCCTTACGCTGGGAGACGACTTTGGATATGATGGAATCACAGGACCAAATTACTGGGGCGAGAGATATCGAGAATGCGCAGGAAAGCACCAGAGTCCTATCAATATCAACATTCTGCATGTTAAACATATAATTCTACCAGATATAGAGTTCATCGGCTTTGATGACTCGATCGACGATGTACACGTCACCAACAATGGACACACAGTATTAATTGAAGTGAAGAATGAACCTCATCCGAGGGTTCGCGGTGGGCCTTTAGATGGTGATTACATCTTCAGCCAGATGCATTTCCACTGGGGAGATAACGACACGTTTGGCTCTGAAGATACTATCAATCATCGGAGTTTTCCTATGGAATTGCACATGGTGTTCTTCAAAGAAGAGTATGGAACCCCAACCAACGCAGTGAAGCATCACGATGGATTAACAGTGCTAGCATTTTTTTACGAGTTGGACCGTCATCAGCATCCAGCTTACGATGATATAACATCCGCATTAGCGAATGTAACCGAACCCTATACTTCCGTGGCGATGAGCCATCCCTTTTCATTCCTCAACATTTTGCCTTACGATTTGAAGAGGTACTTTACCTACAAGGGCTCATTGACAACTCCGCCTTGTTCTGAAGTTGTCATTTGGTTGGATTTCGAGCAGCCTATTAGATTAACACACGATCAGATAGAAACGTTTCGTGAACTCAAGTCTGCACACGGAAAGATCACTCATAATTTCCGACCAATACAGCCAATCGGTGACAGAATTGTATTTTATAACATTGACAACAGCTACTTTAGTTACAACACTATAGATGAAGATGAAGAGGAACATAAAGACACGCATCCAAAACGAAAGAAGCATCGCAATAATGCACAAAATATATTAAGCAATTATACTTTAATTTTATGCAATGTCTTGCTTTTAGTATATGGTATTCACTTAAGTCCATTAATTTCAGAGGTCTCTTTCACGACTGATTTCTAA
Protein
MRSVCLVLLTLGDDFGYDGITGPNYWGERYRECAGKHQSPININILHVKHIILPDIEFIGFDDSIDDVHVTNNGHTVLIEVKNEPHPRVRGGPLDGDYIFSQMHFHWGDNDTFGSEDTINHRSFPMELHMVFFKEEYGTPTNAVKHHDGLTVLAFFYELDRHQHPAYDDITSALANVTEPYTSVAMSHPFSFLNILPYDLKRYFTYKGSLTTPPCSEVVIWLDFEQPIRLTHDQIETFRELKSAHGKITHNFRPIQPIGDRIVFYNIDNSYFSYNTIDEDEEEHKDTHPKRKKHRNNAQNILSNYTLILCNVLLLVYGIHLSPLISEVSFTTDF
Summary
Similarity
Belongs to the alpha-carbonic anhydrase family.
Uniprot
A0A2A4JJ54
A0A194QJS8
A0A0N0PAX3
A0A3S2LHQ5
A0A212EZ15
A0A2H1WG46
+ More
W5JG76 A0A182NH68 A0A2M3ZL28 A0A182FG20 A0A182RAY9 A0A1W7R5I4 A0A2M3Z564 A0A182PH89 A0A2M3ZKR5 A0A2M4BSS0 A0A023EQM0 A0A2M4BSF3 A0A2M4BSP1 A0A2M4ADY0 A0A023EQ37 A0A182ME06 Q7QJJ8 A0A182GPB3 A0A182X3J9 A0A1Q3FRE4 A0A182KRJ8 A0A1Q3FRR6 A0A1Q3FRJ5 A0A1Q3FR93 A0A1Q3FRI3 A0A182I3H1 C5ML19 A0A182QLY7 A0A084VCF1 A0A182IYA9 A0A182UU55 A0A182TST6 A0A2M4BTJ4 A0A2M4BUG2 A0A2M4AEI9 A0A182WJ26 Q17AD2 A0A2P8YTE9 B0WW71 A0A182XZ22 A0A182JTX4 A0A336KUN5 A0A1J1J1U8 A0A1B6CME5 A0A1B0CVX3 A0A1W4WKS9 A0A2J7QNX0 A0A2J7QNW5 A0A026VYY7 A0A1W4W9H0 A0A1B6JW96 A0A0C9RJN0 A0A3L8DNH1 A0A069DRW8 E2AME3 A0A0A1WV76 A0A0A1WZZ4 A0A232F4S8 K7J542 A0A151IFZ8 A0A088ATP0 A0A2S2QU51 A0A2H8TD49 M1TA57 W8C6Y4 A0A195FEJ3 E9ILF3 J9K0P1 A0A034VPX0 B4IWN5 A0A195DXS2 A0A158NH74 B4LEI5 A0A195B7N9 N6TUS2 A0A0M9AD54 A0A2A3E5W9 A0A0K8VXF9 Q2M0R9 F4WZM6 B4GR68 D0QWS9 B4KZ41 B3M7V5 A0A3B0J7V3 B4HFJ8 B3NH77 B4N3T1 A0A1I8N8W0 T1PIC4 Q9VTU8 B4QR68
W5JG76 A0A182NH68 A0A2M3ZL28 A0A182FG20 A0A182RAY9 A0A1W7R5I4 A0A2M3Z564 A0A182PH89 A0A2M3ZKR5 A0A2M4BSS0 A0A023EQM0 A0A2M4BSF3 A0A2M4BSP1 A0A2M4ADY0 A0A023EQ37 A0A182ME06 Q7QJJ8 A0A182GPB3 A0A182X3J9 A0A1Q3FRE4 A0A182KRJ8 A0A1Q3FRR6 A0A1Q3FRJ5 A0A1Q3FR93 A0A1Q3FRI3 A0A182I3H1 C5ML19 A0A182QLY7 A0A084VCF1 A0A182IYA9 A0A182UU55 A0A182TST6 A0A2M4BTJ4 A0A2M4BUG2 A0A2M4AEI9 A0A182WJ26 Q17AD2 A0A2P8YTE9 B0WW71 A0A182XZ22 A0A182JTX4 A0A336KUN5 A0A1J1J1U8 A0A1B6CME5 A0A1B0CVX3 A0A1W4WKS9 A0A2J7QNX0 A0A2J7QNW5 A0A026VYY7 A0A1W4W9H0 A0A1B6JW96 A0A0C9RJN0 A0A3L8DNH1 A0A069DRW8 E2AME3 A0A0A1WV76 A0A0A1WZZ4 A0A232F4S8 K7J542 A0A151IFZ8 A0A088ATP0 A0A2S2QU51 A0A2H8TD49 M1TA57 W8C6Y4 A0A195FEJ3 E9ILF3 J9K0P1 A0A034VPX0 B4IWN5 A0A195DXS2 A0A158NH74 B4LEI5 A0A195B7N9 N6TUS2 A0A0M9AD54 A0A2A3E5W9 A0A0K8VXF9 Q2M0R9 F4WZM6 B4GR68 D0QWS9 B4KZ41 B3M7V5 A0A3B0J7V3 B4HFJ8 B3NH77 B4N3T1 A0A1I8N8W0 T1PIC4 Q9VTU8 B4QR68
Pubmed
26354079
22118469
20920257
23761445
24945155
12364791
+ More
14747013 17210077 26483478 20966253 24438588 17510324 29403074 25244985 24508170 30249741 26334808 20798317 25830018 28648823 20075255 23194342 24495485 21282665 25348373 17994087 21347285 23537049 15632085 21719571 19859648 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249
14747013 17210077 26483478 20966253 24438588 17510324 29403074 25244985 24508170 30249741 26334808 20798317 25830018 28648823 20075255 23194342 24495485 21282665 25348373 17994087 21347285 23537049 15632085 21719571 19859648 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249
EMBL
NWSH01001295
PCG71806.1
KQ459144
KPJ03706.1
KQ461133
KPJ08878.1
+ More
RSAL01000007 RVE54072.1 AGBW02011400 OWR46722.1 ODYU01008439 SOQ52053.1 ADMH02001258 ETN63372.1 GGFM01008367 MBW29118.1 GEHC01001255 JAV46390.1 GGFM01002837 MBW23588.1 GGFM01008368 MBW29119.1 GGFJ01006830 MBW55971.1 GAPW01002213 JAC11385.1 GGFJ01006801 MBW55942.1 GGFJ01006800 MBW55941.1 GGFK01005591 MBW38912.1 GAPW01002415 JAC11183.1 AXCM01004824 AAAB01008807 EAA04474.4 EDO64544.1 JXUM01078110 JXUM01078111 KQ563045 KXJ74574.1 GFDL01004930 JAV30115.1 GFDL01004922 JAV30123.1 GFDL01004858 JAV30187.1 GFDL01004936 JAV30109.1 GFDL01004949 JAV30096.1 APCN01000209 GQ121043 ACS28257.1 AXCN02000890 ATLV01010669 KE524597 KFB35645.1 GGFJ01007256 MBW56397.1 GGFJ01007257 MBW56398.1 GGFK01005874 MBW39195.1 CH477336 EAT43229.1 PYGN01000371 PSN47515.1 DS232139 EDS35903.1 UFQS01000530 UFQT01000530 SSX04650.1 SSX25013.1 CVRI01000067 CRL06421.1 GEDC01022678 JAS14620.1 AJWK01031368 AJWK01031369 NEVH01012562 PNF30278.1 PNF30277.1 KK107681 EZA48074.1 GECU01004267 JAT03440.1 GBYB01013472 JAG83239.1 QOIP01000006 RLU21836.1 GBGD01002443 JAC86446.1 GL440813 EFN65367.1 GBXI01011540 JAD02752.1 GBXI01012511 GBXI01009658 JAD01781.1 JAD04634.1 NNAY01000936 OXU25836.1 AAZX01000348 KQ977793 KYM99677.1 GGMS01012076 MBY81279.1 GFXV01000218 MBW12023.1 JX134494 AGG35950.1 GAMC01008276 GAMC01008275 JAB98279.1 KQ981625 KYN39100.1 GL764074 EFZ18639.1 ABLF02027416 GAKP01013612 GAKP01013611 GAKP01013610 JAC45341.1 CH916366 EDV96261.1 KQ980107 KYN17673.1 ADTU01015190 ADTU01015191 CH940647 EDW69070.1 KQ976574 KYM80260.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72116.1 ENN81276.1 ERL91090.1 KQ435698 KOX80709.1 KZ288357 PBC27098.1 GDHF01033479 GDHF01025490 GDHF01008736 GDHF01004385 JAI18835.1 JAI26824.1 JAI43578.1 JAI47929.1 CH379069 EAL30861.1 GL888480 EGI60301.1 CH479188 EDW40253.1 FJ821035 ACN94775.1 CH933809 EDW17838.1 CH902618 EDV38828.1 OUUW01000002 SPP76273.1 CH480815 EDW41229.1 CH954178 EDV51534.1 CH964095 EDW79286.2 KA648496 AFP63125.1 AE014296 AY095036 AAF49948.1 AAM11364.1 CM000363 CM002912 EDX10198.1 KMY99180.1
RSAL01000007 RVE54072.1 AGBW02011400 OWR46722.1 ODYU01008439 SOQ52053.1 ADMH02001258 ETN63372.1 GGFM01008367 MBW29118.1 GEHC01001255 JAV46390.1 GGFM01002837 MBW23588.1 GGFM01008368 MBW29119.1 GGFJ01006830 MBW55971.1 GAPW01002213 JAC11385.1 GGFJ01006801 MBW55942.1 GGFJ01006800 MBW55941.1 GGFK01005591 MBW38912.1 GAPW01002415 JAC11183.1 AXCM01004824 AAAB01008807 EAA04474.4 EDO64544.1 JXUM01078110 JXUM01078111 KQ563045 KXJ74574.1 GFDL01004930 JAV30115.1 GFDL01004922 JAV30123.1 GFDL01004858 JAV30187.1 GFDL01004936 JAV30109.1 GFDL01004949 JAV30096.1 APCN01000209 GQ121043 ACS28257.1 AXCN02000890 ATLV01010669 KE524597 KFB35645.1 GGFJ01007256 MBW56397.1 GGFJ01007257 MBW56398.1 GGFK01005874 MBW39195.1 CH477336 EAT43229.1 PYGN01000371 PSN47515.1 DS232139 EDS35903.1 UFQS01000530 UFQT01000530 SSX04650.1 SSX25013.1 CVRI01000067 CRL06421.1 GEDC01022678 JAS14620.1 AJWK01031368 AJWK01031369 NEVH01012562 PNF30278.1 PNF30277.1 KK107681 EZA48074.1 GECU01004267 JAT03440.1 GBYB01013472 JAG83239.1 QOIP01000006 RLU21836.1 GBGD01002443 JAC86446.1 GL440813 EFN65367.1 GBXI01011540 JAD02752.1 GBXI01012511 GBXI01009658 JAD01781.1 JAD04634.1 NNAY01000936 OXU25836.1 AAZX01000348 KQ977793 KYM99677.1 GGMS01012076 MBY81279.1 GFXV01000218 MBW12023.1 JX134494 AGG35950.1 GAMC01008276 GAMC01008275 JAB98279.1 KQ981625 KYN39100.1 GL764074 EFZ18639.1 ABLF02027416 GAKP01013612 GAKP01013611 GAKP01013610 JAC45341.1 CH916366 EDV96261.1 KQ980107 KYN17673.1 ADTU01015190 ADTU01015191 CH940647 EDW69070.1 KQ976574 KYM80260.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72116.1 ENN81276.1 ERL91090.1 KQ435698 KOX80709.1 KZ288357 PBC27098.1 GDHF01033479 GDHF01025490 GDHF01008736 GDHF01004385 JAI18835.1 JAI26824.1 JAI43578.1 JAI47929.1 CH379069 EAL30861.1 GL888480 EGI60301.1 CH479188 EDW40253.1 FJ821035 ACN94775.1 CH933809 EDW17838.1 CH902618 EDV38828.1 OUUW01000002 SPP76273.1 CH480815 EDW41229.1 CH954178 EDV51534.1 CH964095 EDW79286.2 KA648496 AFP63125.1 AE014296 AY095036 AAF49948.1 AAM11364.1 CM000363 CM002912 EDX10198.1 KMY99180.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000000673
+ More
UP000075884 UP000069272 UP000075900 UP000075885 UP000075883 UP000007062 UP000069940 UP000249989 UP000076407 UP000075882 UP000075840 UP000075886 UP000030765 UP000075880 UP000075903 UP000075902 UP000075920 UP000008820 UP000245037 UP000002320 UP000076408 UP000075881 UP000183832 UP000092461 UP000192223 UP000235965 UP000053097 UP000279307 UP000000311 UP000215335 UP000002358 UP000078542 UP000005203 UP000078541 UP000007819 UP000001070 UP000078492 UP000005205 UP000008792 UP000078540 UP000019118 UP000030742 UP000053105 UP000242457 UP000001819 UP000007755 UP000008744 UP000009192 UP000007801 UP000268350 UP000001292 UP000008711 UP000007798 UP000095301 UP000000803 UP000000304
UP000075884 UP000069272 UP000075900 UP000075885 UP000075883 UP000007062 UP000069940 UP000249989 UP000076407 UP000075882 UP000075840 UP000075886 UP000030765 UP000075880 UP000075903 UP000075902 UP000075920 UP000008820 UP000245037 UP000002320 UP000076408 UP000075881 UP000183832 UP000092461 UP000192223 UP000235965 UP000053097 UP000279307 UP000000311 UP000215335 UP000002358 UP000078542 UP000005203 UP000078541 UP000007819 UP000001070 UP000078492 UP000005205 UP000008792 UP000078540 UP000019118 UP000030742 UP000053105 UP000242457 UP000001819 UP000007755 UP000008744 UP000009192 UP000007801 UP000268350 UP000001292 UP000008711 UP000007798 UP000095301 UP000000803 UP000000304
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JJ54
A0A194QJS8
A0A0N0PAX3
A0A3S2LHQ5
A0A212EZ15
A0A2H1WG46
+ More
W5JG76 A0A182NH68 A0A2M3ZL28 A0A182FG20 A0A182RAY9 A0A1W7R5I4 A0A2M3Z564 A0A182PH89 A0A2M3ZKR5 A0A2M4BSS0 A0A023EQM0 A0A2M4BSF3 A0A2M4BSP1 A0A2M4ADY0 A0A023EQ37 A0A182ME06 Q7QJJ8 A0A182GPB3 A0A182X3J9 A0A1Q3FRE4 A0A182KRJ8 A0A1Q3FRR6 A0A1Q3FRJ5 A0A1Q3FR93 A0A1Q3FRI3 A0A182I3H1 C5ML19 A0A182QLY7 A0A084VCF1 A0A182IYA9 A0A182UU55 A0A182TST6 A0A2M4BTJ4 A0A2M4BUG2 A0A2M4AEI9 A0A182WJ26 Q17AD2 A0A2P8YTE9 B0WW71 A0A182XZ22 A0A182JTX4 A0A336KUN5 A0A1J1J1U8 A0A1B6CME5 A0A1B0CVX3 A0A1W4WKS9 A0A2J7QNX0 A0A2J7QNW5 A0A026VYY7 A0A1W4W9H0 A0A1B6JW96 A0A0C9RJN0 A0A3L8DNH1 A0A069DRW8 E2AME3 A0A0A1WV76 A0A0A1WZZ4 A0A232F4S8 K7J542 A0A151IFZ8 A0A088ATP0 A0A2S2QU51 A0A2H8TD49 M1TA57 W8C6Y4 A0A195FEJ3 E9ILF3 J9K0P1 A0A034VPX0 B4IWN5 A0A195DXS2 A0A158NH74 B4LEI5 A0A195B7N9 N6TUS2 A0A0M9AD54 A0A2A3E5W9 A0A0K8VXF9 Q2M0R9 F4WZM6 B4GR68 D0QWS9 B4KZ41 B3M7V5 A0A3B0J7V3 B4HFJ8 B3NH77 B4N3T1 A0A1I8N8W0 T1PIC4 Q9VTU8 B4QR68
W5JG76 A0A182NH68 A0A2M3ZL28 A0A182FG20 A0A182RAY9 A0A1W7R5I4 A0A2M3Z564 A0A182PH89 A0A2M3ZKR5 A0A2M4BSS0 A0A023EQM0 A0A2M4BSF3 A0A2M4BSP1 A0A2M4ADY0 A0A023EQ37 A0A182ME06 Q7QJJ8 A0A182GPB3 A0A182X3J9 A0A1Q3FRE4 A0A182KRJ8 A0A1Q3FRR6 A0A1Q3FRJ5 A0A1Q3FR93 A0A1Q3FRI3 A0A182I3H1 C5ML19 A0A182QLY7 A0A084VCF1 A0A182IYA9 A0A182UU55 A0A182TST6 A0A2M4BTJ4 A0A2M4BUG2 A0A2M4AEI9 A0A182WJ26 Q17AD2 A0A2P8YTE9 B0WW71 A0A182XZ22 A0A182JTX4 A0A336KUN5 A0A1J1J1U8 A0A1B6CME5 A0A1B0CVX3 A0A1W4WKS9 A0A2J7QNX0 A0A2J7QNW5 A0A026VYY7 A0A1W4W9H0 A0A1B6JW96 A0A0C9RJN0 A0A3L8DNH1 A0A069DRW8 E2AME3 A0A0A1WV76 A0A0A1WZZ4 A0A232F4S8 K7J542 A0A151IFZ8 A0A088ATP0 A0A2S2QU51 A0A2H8TD49 M1TA57 W8C6Y4 A0A195FEJ3 E9ILF3 J9K0P1 A0A034VPX0 B4IWN5 A0A195DXS2 A0A158NH74 B4LEI5 A0A195B7N9 N6TUS2 A0A0M9AD54 A0A2A3E5W9 A0A0K8VXF9 Q2M0R9 F4WZM6 B4GR68 D0QWS9 B4KZ41 B3M7V5 A0A3B0J7V3 B4HFJ8 B3NH77 B4N3T1 A0A1I8N8W0 T1PIC4 Q9VTU8 B4QR68
PDB
4PXX
E-value=1.41433e-34,
Score=365
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 12,
Likelihood: 0.519236
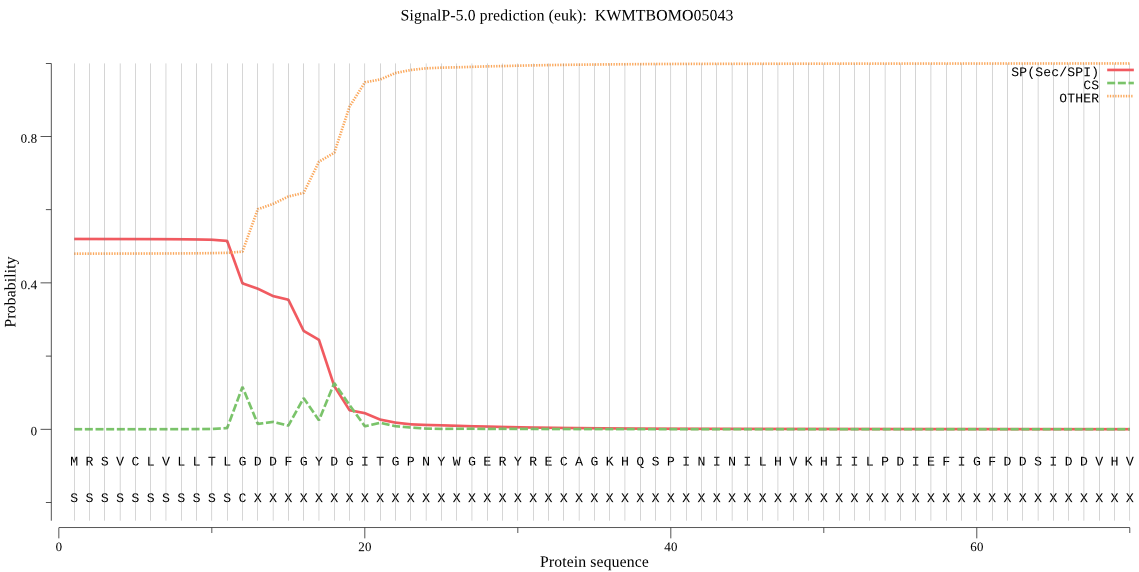
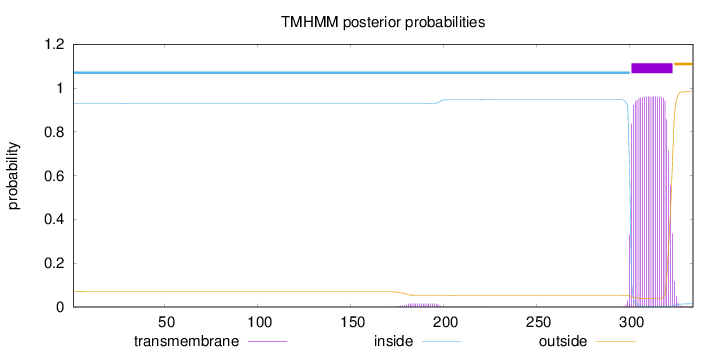
Length:
334
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.42222
Exp number, first 60 AAs:
0.02768
Total prob of N-in:
0.92891
inside
1 - 300
TMhelix
301 - 323
outside
324 - 334
Population Genetic Test Statistics
Pi
250.606385
Theta
202.727419
Tajima's D
0.797574
CLR
0.264897
CSRT
0.604269786510674
Interpretation
Uncertain
