Pre Gene Modal
BGIBMGA012532
Annotation
PREDICTED:_zinc_finger_protein_830_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.907
Sequence
CDS
ATGGCACTCAGGTTGCAAATCGAAAAGAAGAAAGCTCAAGAAGAAATGCGTCGTTTGATGGCAGAAAGGAAGAAGAAGGATGTCAAACGAGTAAAGTTAGATCATCCTCTCGCAAGATATAATAATGTGGGACATCTCACTTGTTTGCTTTGCACTTCCGTCGTGAGATCGGAAAATGTTTGGCAAGTACATCTCAATAGTAAACAACATCGAGAAAATATTGAAAATGCTAAGAAACTAAAGGAACTGACGAATAATTTCACTGTAGTCAACAAAGTTAAACACAAGAGTTCCAGTCCTCCAAAAGATGCACCTCCCGAAAAAAAGATAAAAGGAATATTAAAAAATGTAGGCACCGTGCCAGTGATAACAAAACCCAAAAATGATGTCCCAAGTGTGGTAGAATTTCACAATGAAGAAATTAAAAGAACTCCACTTAATCCTGTTGTAATAACAGAAAAAGTTAATGAACCTGTACAGACTACAGATCAAAAAGAAAAAGAGGAACCTGCAAGTGATGTACCAATACCAGAAGGTTTTTTTGATGACCCAGTTTTAGATGCAAAGGTTCGTAAGATTGAATATAAAGATCCAATTGAAGAGGAATGGGAAAAATTCAGAAAAGAAATAAAAGAAGAAACTAGTGCATCAGCTGAAATTATAGCAGGTGAACAAGAAGAAGCCACTGCAGAGCGACAAATAGAAGAAATCGACGAACAAATTAGAAAATGGTCCCGAGTCCTGGATTTGGAATTACAAAAAGAAGAGACGAAAAAGAAAATACACGAAATGGACCAAATGAGCTTAGACGAAAATTGTGAATCAGAAAACGAAGATGATATAGACGAATTTTTAGATTGGAGGTCTAAAAAATCGTTTACGTAA
Protein
MALRLQIEKKKAQEEMRRLMAERKKKDVKRVKLDHPLARYNNVGHLTCLLCTSVVRSENVWQVHLNSKQHRENIENAKKLKELTNNFTVVNKVKHKSSSPPKDAPPEKKIKGILKNVGTVPVITKPKNDVPSVVEFHNEEIKRTPLNPVVITEKVNEPVQTTDQKEKEEPASDVPIPEGFFDDPVLDAKVRKIEYKDPIEEEWEKFRKEIKEETSASAEIIAGEQEEATAERQIEEIDEQIRKWSRVLDLELQKEETKKKIHEMDQMSLDENCESENEDDIDEFLDWRSKKSFT
Summary
Uniprot
H9JSM1
A0A2A4IT60
A0A2H1WP75
A0A0N0PAX7
A0A212EYZ1
S4PX00
+ More
A0A194QJT6 A0A0L7L7E9 A0A0C9R6D6 A0A0T6AYG5 A0A310S8U8 A0A1W4XCG2 A0A158P3T8 A0A067QKX1 A0A1L8DA72 D6W8Z6 A0A195FJR7 A0A1Y1NFL8 A0A195DZQ8 A0A026WBR1 A0A151WF45 A0A3L8DRY4 A0A088ADX4 A0A2J7QJ74 K7J233 E9IK22 Q16M83 A0A1S4FVY8 A0A2J7PPE1 A0A1Y1NG92 A0A195AT12 F4WKZ9 A0A182QET8 A0A195CP22 A0A2M3Z9V3 A0A2J7Q561 A0A2M3ZIF1 A0A2A3E462 A0A182H520 A0A2M3ZIH5 A0A0J7K3S8 J3JXK5 A0A2M4CRF2 N6TH90 A0A2M4BSL0 A0A2M4CRK3 A0A2M4BSN6 A0A2M4BSB3 A0A1B6FUH5 B0WL11 A0A0A9Y6Q5 A0A1B6LLH5 A0A1B0CMF2 A0A2P8ZCN4 A0A1B6K007 A0A1Q3F1H6 E0VB24 A0A0P4VLA4 A0A1Q3F1A3 T1I939 A0A224XTM1 A0A0V0GBI3 A0A1Q3F3D9 R4G4Q9 A0A182YG76 A0A023F7V6 A0A069DRI3 U5EY19 A0A2M3ZJ01 A0A336KRZ7 A0A336KS75 Q7Q7N4 E9GGP7 A0A182VPP3 A0A182FP93 A0A232ESP0 A0A182TAZ7 A0A084W7T3 A0A182XDP2 A0A182REC9 A0A0L7RGT8 A0A182IQX6 A0A1J1HSE3 A0A023EP51 A0A1E1XGY5 G3MKX2 A0A0C9RUN9 A0A023GKF3 A0A0N8A2W2 A0A0K8TR77 A0A0P5SV75 A0A2S2PDS6 A0A3Q0IZW1 E2BFQ6 A0A2H8U1P9 A0A0P6AKF9 A0A131YAT7 X1X0S2
A0A194QJT6 A0A0L7L7E9 A0A0C9R6D6 A0A0T6AYG5 A0A310S8U8 A0A1W4XCG2 A0A158P3T8 A0A067QKX1 A0A1L8DA72 D6W8Z6 A0A195FJR7 A0A1Y1NFL8 A0A195DZQ8 A0A026WBR1 A0A151WF45 A0A3L8DRY4 A0A088ADX4 A0A2J7QJ74 K7J233 E9IK22 Q16M83 A0A1S4FVY8 A0A2J7PPE1 A0A1Y1NG92 A0A195AT12 F4WKZ9 A0A182QET8 A0A195CP22 A0A2M3Z9V3 A0A2J7Q561 A0A2M3ZIF1 A0A2A3E462 A0A182H520 A0A2M3ZIH5 A0A0J7K3S8 J3JXK5 A0A2M4CRF2 N6TH90 A0A2M4BSL0 A0A2M4CRK3 A0A2M4BSN6 A0A2M4BSB3 A0A1B6FUH5 B0WL11 A0A0A9Y6Q5 A0A1B6LLH5 A0A1B0CMF2 A0A2P8ZCN4 A0A1B6K007 A0A1Q3F1H6 E0VB24 A0A0P4VLA4 A0A1Q3F1A3 T1I939 A0A224XTM1 A0A0V0GBI3 A0A1Q3F3D9 R4G4Q9 A0A182YG76 A0A023F7V6 A0A069DRI3 U5EY19 A0A2M3ZJ01 A0A336KRZ7 A0A336KS75 Q7Q7N4 E9GGP7 A0A182VPP3 A0A182FP93 A0A232ESP0 A0A182TAZ7 A0A084W7T3 A0A182XDP2 A0A182REC9 A0A0L7RGT8 A0A182IQX6 A0A1J1HSE3 A0A023EP51 A0A1E1XGY5 G3MKX2 A0A0C9RUN9 A0A023GKF3 A0A0N8A2W2 A0A0K8TR77 A0A0P5SV75 A0A2S2PDS6 A0A3Q0IZW1 E2BFQ6 A0A2H8U1P9 A0A0P6AKF9 A0A131YAT7 X1X0S2
Pubmed
19121390
26354079
22118469
23622113
26227816
21347285
+ More
24845553 18362917 19820115 28004739 24508170 30249741 20075255 21282665 17510324 21719571 26483478 22516182 23537049 25401762 26823975 29403074 20566863 27129103 25244985 25474469 26334808 12364791 14747013 17210077 21292972 28648823 24438588 24945155 28503490 22216098 26131772 26369729 20798317
24845553 18362917 19820115 28004739 24508170 30249741 20075255 21282665 17510324 21719571 26483478 22516182 23537049 25401762 26823975 29403074 20566863 27129103 25244985 25474469 26334808 12364791 14747013 17210077 21292972 28648823 24438588 24945155 28503490 22216098 26131772 26369729 20798317
EMBL
BABH01032184
BABH01032185
NWSH01008411
PCG62568.1
ODYU01010043
SOQ54870.1
+ More
KQ461133 KPJ08888.1 AGBW02011400 OWR46713.1 GAIX01005903 JAA86657.1 KQ459144 KPJ03716.1 JTDY01002544 KOB71211.1 GBYB01002316 JAG72083.1 LJIG01022517 KRT80156.1 KQ769818 OAD52801.1 ADTU01008450 KK853234 KDR09596.1 GFDF01010726 JAV03358.1 KQ971312 EEZ98228.2 KQ981523 KYN40502.1 GEZM01003800 JAV96559.1 KQ980044 KYN18109.1 KK107293 EZA53403.1 KQ983227 KYQ46442.1 QOIP01000005 RLU23177.1 NEVH01013559 PNF28637.1 GL763924 EFZ19017.1 CH477873 EAT35438.1 NEVH01023278 PNF18171.1 GEZM01003801 JAV96558.1 KQ976746 KYM75358.1 GL888206 EGI65241.1 AXCN02001594 KQ977513 KYN02232.1 GGFM01004509 MBW25260.1 NEVH01018370 PNF23721.1 GGFM01007550 MBW28301.1 KZ288416 PBC26046.1 JXUM01110644 KQ565431 KXJ70980.1 GGFM01007593 MBW28344.1 LBMM01015359 KMQ84841.1 BT127974 KB632345 AEE62936.1 ERL93103.1 GGFL01003732 MBW67910.1 APGK01026786 APGK01026787 KB740642 ENN79799.1 GGFJ01006908 MBW56049.1 GGFL01003731 MBW67909.1 GGFJ01006954 MBW56095.1 GGFJ01006836 MBW55977.1 GECZ01015897 JAS53872.1 DS231979 EDS30174.1 GBHO01016836 GBRD01016789 GDHC01007670 JAG26768.1 JAG49038.1 JAQ10959.1 GEBQ01015386 JAT24591.1 AJWK01018646 PYGN01000103 PSN54259.1 GECU01002924 JAT04783.1 GFDL01013641 JAV21404.1 DS235021 EEB10580.1 GDKW01001410 JAI55185.1 GFDL01013713 JAV21332.1 ACPB03021415 GFTR01004554 JAW11872.1 GECL01000635 JAP05489.1 GFDL01012974 JAV22071.1 GAHY01001135 JAA76375.1 GBBI01001382 JAC17330.1 GBGD01002369 JAC86520.1 GANO01000604 JAB59267.1 GGFM01007694 MBW28445.1 UFQS01000413 UFQT01000413 SSX03720.1 SSX24085.1 UFQS01000418 UFQT01000418 SSX03790.1 SSX24155.1 AAAB01008952 EAA10595.5 GL732543 EFX81429.1 NNAY01002419 OXU21316.1 ATLV01021296 KE525316 KFB46277.1 KQ414596 KOC69906.1 CVRI01000020 CRK90925.1 GAPW01003349 JAC10249.1 GFAC01000690 JAT98498.1 JO842523 AEO34140.1 GBZX01001569 JAG91171.1 GBBM01001905 JAC33513.1 GDIP01187841 JAJ35561.1 GDAI01000972 JAI16631.1 GDIP01134813 GDIQ01046533 LRGB01002101 JAL68901.1 JAN48204.1 KZS09214.1 GGMR01014991 MBY27610.1 GL448039 EFN85444.1 GFXV01007563 MBW19368.1 GDIP01028394 JAM75321.1 GEFM01000600 JAP75196.1 ABLF02018087
KQ461133 KPJ08888.1 AGBW02011400 OWR46713.1 GAIX01005903 JAA86657.1 KQ459144 KPJ03716.1 JTDY01002544 KOB71211.1 GBYB01002316 JAG72083.1 LJIG01022517 KRT80156.1 KQ769818 OAD52801.1 ADTU01008450 KK853234 KDR09596.1 GFDF01010726 JAV03358.1 KQ971312 EEZ98228.2 KQ981523 KYN40502.1 GEZM01003800 JAV96559.1 KQ980044 KYN18109.1 KK107293 EZA53403.1 KQ983227 KYQ46442.1 QOIP01000005 RLU23177.1 NEVH01013559 PNF28637.1 GL763924 EFZ19017.1 CH477873 EAT35438.1 NEVH01023278 PNF18171.1 GEZM01003801 JAV96558.1 KQ976746 KYM75358.1 GL888206 EGI65241.1 AXCN02001594 KQ977513 KYN02232.1 GGFM01004509 MBW25260.1 NEVH01018370 PNF23721.1 GGFM01007550 MBW28301.1 KZ288416 PBC26046.1 JXUM01110644 KQ565431 KXJ70980.1 GGFM01007593 MBW28344.1 LBMM01015359 KMQ84841.1 BT127974 KB632345 AEE62936.1 ERL93103.1 GGFL01003732 MBW67910.1 APGK01026786 APGK01026787 KB740642 ENN79799.1 GGFJ01006908 MBW56049.1 GGFL01003731 MBW67909.1 GGFJ01006954 MBW56095.1 GGFJ01006836 MBW55977.1 GECZ01015897 JAS53872.1 DS231979 EDS30174.1 GBHO01016836 GBRD01016789 GDHC01007670 JAG26768.1 JAG49038.1 JAQ10959.1 GEBQ01015386 JAT24591.1 AJWK01018646 PYGN01000103 PSN54259.1 GECU01002924 JAT04783.1 GFDL01013641 JAV21404.1 DS235021 EEB10580.1 GDKW01001410 JAI55185.1 GFDL01013713 JAV21332.1 ACPB03021415 GFTR01004554 JAW11872.1 GECL01000635 JAP05489.1 GFDL01012974 JAV22071.1 GAHY01001135 JAA76375.1 GBBI01001382 JAC17330.1 GBGD01002369 JAC86520.1 GANO01000604 JAB59267.1 GGFM01007694 MBW28445.1 UFQS01000413 UFQT01000413 SSX03720.1 SSX24085.1 UFQS01000418 UFQT01000418 SSX03790.1 SSX24155.1 AAAB01008952 EAA10595.5 GL732543 EFX81429.1 NNAY01002419 OXU21316.1 ATLV01021296 KE525316 KFB46277.1 KQ414596 KOC69906.1 CVRI01000020 CRK90925.1 GAPW01003349 JAC10249.1 GFAC01000690 JAT98498.1 JO842523 AEO34140.1 GBZX01001569 JAG91171.1 GBBM01001905 JAC33513.1 GDIP01187841 JAJ35561.1 GDAI01000972 JAI16631.1 GDIP01134813 GDIQ01046533 LRGB01002101 JAL68901.1 JAN48204.1 KZS09214.1 GGMR01014991 MBY27610.1 GL448039 EFN85444.1 GFXV01007563 MBW19368.1 GDIP01028394 JAM75321.1 GEFM01000600 JAP75196.1 ABLF02018087
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000192223 UP000005205 UP000027135 UP000007266 UP000078541 UP000078492 UP000053097 UP000075809 UP000279307 UP000005203 UP000235965 UP000002358 UP000008820 UP000078540 UP000007755 UP000075886 UP000078542 UP000242457 UP000069940 UP000249989 UP000036403 UP000030742 UP000019118 UP000002320 UP000092461 UP000245037 UP000009046 UP000015103 UP000076408 UP000007062 UP000000305 UP000075920 UP000069272 UP000215335 UP000075901 UP000030765 UP000076407 UP000075900 UP000053825 UP000075880 UP000183832 UP000076858 UP000079169 UP000008237 UP000007819
UP000192223 UP000005205 UP000027135 UP000007266 UP000078541 UP000078492 UP000053097 UP000075809 UP000279307 UP000005203 UP000235965 UP000002358 UP000008820 UP000078540 UP000007755 UP000075886 UP000078542 UP000242457 UP000069940 UP000249989 UP000036403 UP000030742 UP000019118 UP000002320 UP000092461 UP000245037 UP000009046 UP000015103 UP000076408 UP000007062 UP000000305 UP000075920 UP000069272 UP000215335 UP000075901 UP000030765 UP000076407 UP000075900 UP000053825 UP000075880 UP000183832 UP000076858 UP000079169 UP000008237 UP000007819
PRIDE
Interpro
IPR036236
Znf_C2H2_sf
+ More
IPR019538 PSMD5
IPR016024 ARM-type_fold
IPR022755 Znf_C2H2_jaz
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR040050 ZNF830
IPR011989 ARM-like
IPR015424 PyrdxlP-dep_Trfase
IPR000192 Aminotrans_V_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR022278 Pser_aminoTfrase
IPR019538 PSMD5
IPR016024 ARM-type_fold
IPR022755 Znf_C2H2_jaz
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR040050 ZNF830
IPR011989 ARM-like
IPR015424 PyrdxlP-dep_Trfase
IPR000192 Aminotrans_V_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR022278 Pser_aminoTfrase
Gene 3D
ProteinModelPortal
H9JSM1
A0A2A4IT60
A0A2H1WP75
A0A0N0PAX7
A0A212EYZ1
S4PX00
+ More
A0A194QJT6 A0A0L7L7E9 A0A0C9R6D6 A0A0T6AYG5 A0A310S8U8 A0A1W4XCG2 A0A158P3T8 A0A067QKX1 A0A1L8DA72 D6W8Z6 A0A195FJR7 A0A1Y1NFL8 A0A195DZQ8 A0A026WBR1 A0A151WF45 A0A3L8DRY4 A0A088ADX4 A0A2J7QJ74 K7J233 E9IK22 Q16M83 A0A1S4FVY8 A0A2J7PPE1 A0A1Y1NG92 A0A195AT12 F4WKZ9 A0A182QET8 A0A195CP22 A0A2M3Z9V3 A0A2J7Q561 A0A2M3ZIF1 A0A2A3E462 A0A182H520 A0A2M3ZIH5 A0A0J7K3S8 J3JXK5 A0A2M4CRF2 N6TH90 A0A2M4BSL0 A0A2M4CRK3 A0A2M4BSN6 A0A2M4BSB3 A0A1B6FUH5 B0WL11 A0A0A9Y6Q5 A0A1B6LLH5 A0A1B0CMF2 A0A2P8ZCN4 A0A1B6K007 A0A1Q3F1H6 E0VB24 A0A0P4VLA4 A0A1Q3F1A3 T1I939 A0A224XTM1 A0A0V0GBI3 A0A1Q3F3D9 R4G4Q9 A0A182YG76 A0A023F7V6 A0A069DRI3 U5EY19 A0A2M3ZJ01 A0A336KRZ7 A0A336KS75 Q7Q7N4 E9GGP7 A0A182VPP3 A0A182FP93 A0A232ESP0 A0A182TAZ7 A0A084W7T3 A0A182XDP2 A0A182REC9 A0A0L7RGT8 A0A182IQX6 A0A1J1HSE3 A0A023EP51 A0A1E1XGY5 G3MKX2 A0A0C9RUN9 A0A023GKF3 A0A0N8A2W2 A0A0K8TR77 A0A0P5SV75 A0A2S2PDS6 A0A3Q0IZW1 E2BFQ6 A0A2H8U1P9 A0A0P6AKF9 A0A131YAT7 X1X0S2
A0A194QJT6 A0A0L7L7E9 A0A0C9R6D6 A0A0T6AYG5 A0A310S8U8 A0A1W4XCG2 A0A158P3T8 A0A067QKX1 A0A1L8DA72 D6W8Z6 A0A195FJR7 A0A1Y1NFL8 A0A195DZQ8 A0A026WBR1 A0A151WF45 A0A3L8DRY4 A0A088ADX4 A0A2J7QJ74 K7J233 E9IK22 Q16M83 A0A1S4FVY8 A0A2J7PPE1 A0A1Y1NG92 A0A195AT12 F4WKZ9 A0A182QET8 A0A195CP22 A0A2M3Z9V3 A0A2J7Q561 A0A2M3ZIF1 A0A2A3E462 A0A182H520 A0A2M3ZIH5 A0A0J7K3S8 J3JXK5 A0A2M4CRF2 N6TH90 A0A2M4BSL0 A0A2M4CRK3 A0A2M4BSN6 A0A2M4BSB3 A0A1B6FUH5 B0WL11 A0A0A9Y6Q5 A0A1B6LLH5 A0A1B0CMF2 A0A2P8ZCN4 A0A1B6K007 A0A1Q3F1H6 E0VB24 A0A0P4VLA4 A0A1Q3F1A3 T1I939 A0A224XTM1 A0A0V0GBI3 A0A1Q3F3D9 R4G4Q9 A0A182YG76 A0A023F7V6 A0A069DRI3 U5EY19 A0A2M3ZJ01 A0A336KRZ7 A0A336KS75 Q7Q7N4 E9GGP7 A0A182VPP3 A0A182FP93 A0A232ESP0 A0A182TAZ7 A0A084W7T3 A0A182XDP2 A0A182REC9 A0A0L7RGT8 A0A182IQX6 A0A1J1HSE3 A0A023EP51 A0A1E1XGY5 G3MKX2 A0A0C9RUN9 A0A023GKF3 A0A0N8A2W2 A0A0K8TR77 A0A0P5SV75 A0A2S2PDS6 A0A3Q0IZW1 E2BFQ6 A0A2H8U1P9 A0A0P6AKF9 A0A131YAT7 X1X0S2
Ontologies
GO
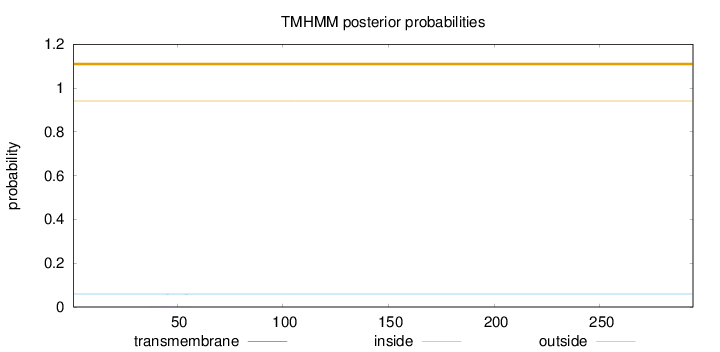
Topology
Length:
294
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00384
Exp number, first 60 AAs:
0.00376
Total prob of N-in:
0.05920
outside
1 - 294
Population Genetic Test Statistics
Pi
156.142559
Theta
158.318145
Tajima's D
-0.459028
CLR
0.093279
CSRT
0.248337583120844
Interpretation
Uncertain
