Gene
KWMTBOMO05032 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012571
Annotation
signal_recognition_particle_19_kDa_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.239
Sequence
CDS
ATGGCTAGTATGATAACATCTTGGAATCCCGAAAAGAAGCACAGTGATGTGGAAAGATGGATATGCATTTACCCTGCATATTTGAACAGCAAAAAGACTCTCGCAGAAGGACGCAGACTTCCTAAATCTGTTTGTGTCGAAAACCCTACACATCAAGAAATTAGAGATGTTCTTCTTGCCACTGGGTTACGAGTAGGGGTTGAAAACAAATTGTATTCAAGAGAATGCAGCAAGGAAATGTTGTACAGAGGCAGAATACGCGTTCAAATTAAGAATGATGATGGTGCCCCTGTAAATCCTGAATTTCCTACTAGAGAATCTGTAATGAAATATATTGGTGAATCCATTCCAAAGCTAAAGACAAGACAGAATCGACCTGTGGAACAACAACCTCAACCAACAAATCAGGCAATCAGTAAGACAAAAGGGAAAAAAGGAAGGCGATGA
Protein
MASMITSWNPEKKHSDVERWICIYPAYLNSKKTLAEGRRLPKSVCVENPTHQEIRDVLLATGLRVGVENKLYSRECSKEMLYRGRIRVQIKNDDGAPVNPEFPTRESVMKYIGESIPKLKTRQNRPVEQQPQPTNQAISKTKGKKGRR
Summary
Uniprot
Q1HQ41
A0A1E1WRD3
A0A194QFJ8
H9JSR0
A0A0N1PGW2
S4PYH7
+ More
A0A212EYZ8 A0A2H1WR08 A0A2J7QF43 A0A0A9Z8G3 A0A0T6BG91 A0A1B6D4C1 A0A2P8XF57 A0A0N0BKM3 A0A2A3E7A7 V9IGH9 A0A088ATP6 V5IAW8 A0A2R7VWN8 A0A0L7L6S7 A0A310SHZ7 R4WDL8 D6W9A1 A0A1B6GK49 A0A158NGR7 A0A151XAC3 A0A195DXV4 A0A0L7QLH6 A0A026VX70 A0A1W4WEQ8 A0A151IFL0 A0A224XZ92 E9ILD6 J3JX07 K7IYD2 R4G882 A0A232F8L8 A0A0P4VSK9 A6YPS1 E2B3V1 A0A1B6M517 A0A1Y1MPL5 A0A131XD43 A0A1B0GLN9 A0A1Y1MHT7 A0A0K8W5Y6 A0A0K8TTI0 A0A224Z157 A0A131YW60 L7M309 A0A0L0CCD8 A0A1B0D2K8 A0A154P9T5 A0A0A1XFG0 N6TUS2 A0A034WJ33 A0A195FFC6 A0A023GF27 A0A023FFJ2 A0A087TGC9 T1P8V6 G3MLV9 A0A1L8ECA1 U5EUC3 A0A195B7P9 A0A336MX53 W8CE95 A0A1I8NWH0 A0A023FWK3 F4WZL4 A0A131Y8E8 E1ZV52 A0A1B6GYD5 A0A1L8DVA3 A0A1S3DEK1 A0A0K8RQL2 B4J367 A0A0J7KPV6 A0A0M4E9J5 B4HV51 B4KZW8 T1DJV8 A0A1W4UGS4 B4PK62 A0A1B6JY16 A0A1B6GPZ7 B4LFC2 A0A1Z5L733 B4QKC9 A0A293M4P5 Q17BN4 A0A1A9X2U7 B5M0L7 B3NFE9 B4MLE4 A0A023EHB4 A0A182GLI4 A0A182PIN3 A0A1A9V594 A0A1E1WWR6
A0A212EYZ8 A0A2H1WR08 A0A2J7QF43 A0A0A9Z8G3 A0A0T6BG91 A0A1B6D4C1 A0A2P8XF57 A0A0N0BKM3 A0A2A3E7A7 V9IGH9 A0A088ATP6 V5IAW8 A0A2R7VWN8 A0A0L7L6S7 A0A310SHZ7 R4WDL8 D6W9A1 A0A1B6GK49 A0A158NGR7 A0A151XAC3 A0A195DXV4 A0A0L7QLH6 A0A026VX70 A0A1W4WEQ8 A0A151IFL0 A0A224XZ92 E9ILD6 J3JX07 K7IYD2 R4G882 A0A232F8L8 A0A0P4VSK9 A6YPS1 E2B3V1 A0A1B6M517 A0A1Y1MPL5 A0A131XD43 A0A1B0GLN9 A0A1Y1MHT7 A0A0K8W5Y6 A0A0K8TTI0 A0A224Z157 A0A131YW60 L7M309 A0A0L0CCD8 A0A1B0D2K8 A0A154P9T5 A0A0A1XFG0 N6TUS2 A0A034WJ33 A0A195FFC6 A0A023GF27 A0A023FFJ2 A0A087TGC9 T1P8V6 G3MLV9 A0A1L8ECA1 U5EUC3 A0A195B7P9 A0A336MX53 W8CE95 A0A1I8NWH0 A0A023FWK3 F4WZL4 A0A131Y8E8 E1ZV52 A0A1B6GYD5 A0A1L8DVA3 A0A1S3DEK1 A0A0K8RQL2 B4J367 A0A0J7KPV6 A0A0M4E9J5 B4HV51 B4KZW8 T1DJV8 A0A1W4UGS4 B4PK62 A0A1B6JY16 A0A1B6GPZ7 B4LFC2 A0A1Z5L733 B4QKC9 A0A293M4P5 Q17BN4 A0A1A9X2U7 B5M0L7 B3NFE9 B4MLE4 A0A023EHB4 A0A182GLI4 A0A182PIN3 A0A1A9V594 A0A1E1WWR6
Pubmed
26354079
19121390
23622113
22118469
25401762
26823975
+ More
29403074 26227816 23691247 18362917 19820115 21347285 24508170 30249741 21282665 22516182 20075255 28648823 27129103 18207082 20798317 28004739 28049606 26369729 28797301 26830274 25576852 26108605 25830018 23537049 25348373 25315136 22216098 24495485 21719571 17994087 24330624 17550304 28528879 22936249 17510324 19166301 24945155 26483478 28503490
29403074 26227816 23691247 18362917 19820115 21347285 24508170 30249741 21282665 22516182 20075255 28648823 27129103 18207082 20798317 28004739 28049606 26369729 28797301 26830274 25576852 26108605 25830018 23537049 25348373 25315136 22216098 24495485 21719571 17994087 24330624 17550304 28528879 22936249 17510324 19166301 24945155 26483478 28503490
EMBL
DQ443211
ABF51300.1
GDQN01001506
JAT89548.1
KQ459144
KPJ03720.1
+ More
BABH01032192 KQ461133 KPJ08892.1 GAIX01003268 JAA89292.1 AGBW02011400 OWR46710.1 ODYU01010043 SOQ54874.1 NEVH01015302 PNF27204.1 GBHO01002875 GBRD01000178 GDHC01014170 GDHC01010509 JAG40729.1 JAG65643.1 JAQ04459.1 JAQ08120.1 LJIG01000582 KRT86358.1 GEDC01025970 GEDC01016778 JAS11328.1 JAS20520.1 PYGN01002371 PSN30640.1 KQ435698 KOX80722.1 KZ288357 PBC27106.1 JR046506 AEY60208.1 GALX01000460 JAB68006.1 KK854101 PTY11311.1 JTDY01002608 KOB71105.1 KQ765373 OAD53960.1 AK417644 BAN20859.1 KQ971312 EEZ98192.1 GECZ01007009 JAS62760.1 ADTU01015173 KQ982339 KYQ57316.1 KQ980107 KYN17656.1 KQ414915 KOC59477.1 KK107681 QOIP01000006 EZA48091.1 RLU21551.1 KQ977793 KYM99664.1 GFTR01002486 JAW13940.1 GL764074 EFZ18605.1 BT127775 AEE62737.1 ACPB03010483 GAHY01001427 JAA76083.1 NNAY01000716 OXU26830.1 GDKW01000466 JAI56129.1 EF639082 ABR27967.1 GL445407 EFN89620.1 GEBQ01008958 JAT31019.1 GEZM01030911 GEZM01030910 GEZM01030909 GEZM01030908 GEZM01030907 GEZM01030906 GEZM01030905 GEZM01030904 GEZM01030903 GEZM01030902 GEZM01030901 GEZM01030898 GEZM01030896 GEZM01030895 GEZM01030894 GEZM01030893 GEZM01030890 GEZM01030889 GEZM01030888 GEZM01030887 GEZM01030886 GEZM01030885 GEZM01030884 GEZM01030883 GEZM01030882 GEZM01030881 GEZM01030880 GEZM01030879 GEZM01030878 GEZM01030877 GEZM01030876 GEZM01030875 JAV85177.1 GEFH01003692 JAP64889.1 AJWK01032579 AJWK01032580 GEZM01030899 GEZM01030897 GEZM01030892 GEZM01030891 JAV85221.1 GDHF01006044 JAI46270.1 GDAI01000363 JAI17240.1 GFPF01011759 MAA22905.1 GEDV01006396 JAP82161.1 GACK01006333 JAA58701.1 JRES01000611 KNC29896.1 AJVK01022918 KQ434851 KZC08612.1 GBXI01004153 JAD10139.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72116.1 ENN81276.1 ERL91090.1 GAKP01004600 JAC54352.1 KQ981625 KYN39088.1 GBBM01002936 JAC32482.1 GBBK01004140 JAC20342.1 KK115093 KFM64168.1 KA644570 AFP59199.1 JO842860 AEO34477.1 GFDG01002543 JAV16256.1 GANO01002384 JAB57487.1 KQ976574 KYM80272.1 UFQS01000200 UFQS01003214 UFQT01000200 UFQT01003214 SSX01240.1 SSX34740.1 GAMC01000586 JAC05970.1 GBBL01002212 JAC25108.1 GL888480 EGI60289.1 GEFM01000297 JAP75499.1 GL434441 EFN74911.1 GECZ01002336 JAS67433.1 GFDF01003741 JAV10343.1 GADI01000431 JAA73377.1 CH916366 EDV96138.1 LBMM01004578 KMQ92269.1 CP012525 ALC43778.1 CH480817 EDW50822.1 CH933809 EDW17980.1 GALA01000031 JAA94821.1 CM000159 EDW93742.1 GECU01003634 JAT04073.1 GECZ01005257 JAS64512.1 CH940647 EDW69220.1 GFJQ02003855 JAW03115.1 CM000363 CM002912 EDX09552.1 KMY98067.1 GFWV01010332 MAA35061.1 CH477319 EAT43679.1 EU930203 ACH56831.1 CH954178 EDV50491.1 CH963847 EDW73402.1 GAPW01005318 JAC08280.1 JXUM01072247 KQ562710 KXJ75286.1 GFAC01007952 JAT91236.1
BABH01032192 KQ461133 KPJ08892.1 GAIX01003268 JAA89292.1 AGBW02011400 OWR46710.1 ODYU01010043 SOQ54874.1 NEVH01015302 PNF27204.1 GBHO01002875 GBRD01000178 GDHC01014170 GDHC01010509 JAG40729.1 JAG65643.1 JAQ04459.1 JAQ08120.1 LJIG01000582 KRT86358.1 GEDC01025970 GEDC01016778 JAS11328.1 JAS20520.1 PYGN01002371 PSN30640.1 KQ435698 KOX80722.1 KZ288357 PBC27106.1 JR046506 AEY60208.1 GALX01000460 JAB68006.1 KK854101 PTY11311.1 JTDY01002608 KOB71105.1 KQ765373 OAD53960.1 AK417644 BAN20859.1 KQ971312 EEZ98192.1 GECZ01007009 JAS62760.1 ADTU01015173 KQ982339 KYQ57316.1 KQ980107 KYN17656.1 KQ414915 KOC59477.1 KK107681 QOIP01000006 EZA48091.1 RLU21551.1 KQ977793 KYM99664.1 GFTR01002486 JAW13940.1 GL764074 EFZ18605.1 BT127775 AEE62737.1 ACPB03010483 GAHY01001427 JAA76083.1 NNAY01000716 OXU26830.1 GDKW01000466 JAI56129.1 EF639082 ABR27967.1 GL445407 EFN89620.1 GEBQ01008958 JAT31019.1 GEZM01030911 GEZM01030910 GEZM01030909 GEZM01030908 GEZM01030907 GEZM01030906 GEZM01030905 GEZM01030904 GEZM01030903 GEZM01030902 GEZM01030901 GEZM01030898 GEZM01030896 GEZM01030895 GEZM01030894 GEZM01030893 GEZM01030890 GEZM01030889 GEZM01030888 GEZM01030887 GEZM01030886 GEZM01030885 GEZM01030884 GEZM01030883 GEZM01030882 GEZM01030881 GEZM01030880 GEZM01030879 GEZM01030878 GEZM01030877 GEZM01030876 GEZM01030875 JAV85177.1 GEFH01003692 JAP64889.1 AJWK01032579 AJWK01032580 GEZM01030899 GEZM01030897 GEZM01030892 GEZM01030891 JAV85221.1 GDHF01006044 JAI46270.1 GDAI01000363 JAI17240.1 GFPF01011759 MAA22905.1 GEDV01006396 JAP82161.1 GACK01006333 JAA58701.1 JRES01000611 KNC29896.1 AJVK01022918 KQ434851 KZC08612.1 GBXI01004153 JAD10139.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72116.1 ENN81276.1 ERL91090.1 GAKP01004600 JAC54352.1 KQ981625 KYN39088.1 GBBM01002936 JAC32482.1 GBBK01004140 JAC20342.1 KK115093 KFM64168.1 KA644570 AFP59199.1 JO842860 AEO34477.1 GFDG01002543 JAV16256.1 GANO01002384 JAB57487.1 KQ976574 KYM80272.1 UFQS01000200 UFQS01003214 UFQT01000200 UFQT01003214 SSX01240.1 SSX34740.1 GAMC01000586 JAC05970.1 GBBL01002212 JAC25108.1 GL888480 EGI60289.1 GEFM01000297 JAP75499.1 GL434441 EFN74911.1 GECZ01002336 JAS67433.1 GFDF01003741 JAV10343.1 GADI01000431 JAA73377.1 CH916366 EDV96138.1 LBMM01004578 KMQ92269.1 CP012525 ALC43778.1 CH480817 EDW50822.1 CH933809 EDW17980.1 GALA01000031 JAA94821.1 CM000159 EDW93742.1 GECU01003634 JAT04073.1 GECZ01005257 JAS64512.1 CH940647 EDW69220.1 GFJQ02003855 JAW03115.1 CM000363 CM002912 EDX09552.1 KMY98067.1 GFWV01010332 MAA35061.1 CH477319 EAT43679.1 EU930203 ACH56831.1 CH954178 EDV50491.1 CH963847 EDW73402.1 GAPW01005318 JAC08280.1 JXUM01072247 KQ562710 KXJ75286.1 GFAC01007952 JAT91236.1
Proteomes
UP000053268
UP000005204
UP000053240
UP000007151
UP000235965
UP000245037
+ More
UP000053105 UP000242457 UP000005203 UP000037510 UP000007266 UP000005205 UP000075809 UP000078492 UP000053825 UP000053097 UP000279307 UP000192223 UP000078542 UP000002358 UP000015103 UP000215335 UP000008237 UP000092461 UP000037069 UP000092462 UP000076502 UP000019118 UP000030742 UP000078541 UP000054359 UP000095301 UP000078540 UP000095300 UP000007755 UP000000311 UP000079169 UP000001070 UP000036403 UP000092553 UP000001292 UP000009192 UP000192221 UP000002282 UP000008792 UP000000304 UP000008820 UP000091820 UP000008711 UP000007798 UP000069940 UP000249989 UP000075885 UP000078200
UP000053105 UP000242457 UP000005203 UP000037510 UP000007266 UP000005205 UP000075809 UP000078492 UP000053825 UP000053097 UP000279307 UP000192223 UP000078542 UP000002358 UP000015103 UP000215335 UP000008237 UP000092461 UP000037069 UP000092462 UP000076502 UP000019118 UP000030742 UP000078541 UP000054359 UP000095301 UP000078540 UP000095300 UP000007755 UP000000311 UP000079169 UP000001070 UP000036403 UP000092553 UP000001292 UP000009192 UP000192221 UP000002282 UP000008792 UP000000304 UP000008820 UP000091820 UP000008711 UP000007798 UP000069940 UP000249989 UP000075885 UP000078200
Interpro
Gene 3D
ProteinModelPortal
Q1HQ41
A0A1E1WRD3
A0A194QFJ8
H9JSR0
A0A0N1PGW2
S4PYH7
+ More
A0A212EYZ8 A0A2H1WR08 A0A2J7QF43 A0A0A9Z8G3 A0A0T6BG91 A0A1B6D4C1 A0A2P8XF57 A0A0N0BKM3 A0A2A3E7A7 V9IGH9 A0A088ATP6 V5IAW8 A0A2R7VWN8 A0A0L7L6S7 A0A310SHZ7 R4WDL8 D6W9A1 A0A1B6GK49 A0A158NGR7 A0A151XAC3 A0A195DXV4 A0A0L7QLH6 A0A026VX70 A0A1W4WEQ8 A0A151IFL0 A0A224XZ92 E9ILD6 J3JX07 K7IYD2 R4G882 A0A232F8L8 A0A0P4VSK9 A6YPS1 E2B3V1 A0A1B6M517 A0A1Y1MPL5 A0A131XD43 A0A1B0GLN9 A0A1Y1MHT7 A0A0K8W5Y6 A0A0K8TTI0 A0A224Z157 A0A131YW60 L7M309 A0A0L0CCD8 A0A1B0D2K8 A0A154P9T5 A0A0A1XFG0 N6TUS2 A0A034WJ33 A0A195FFC6 A0A023GF27 A0A023FFJ2 A0A087TGC9 T1P8V6 G3MLV9 A0A1L8ECA1 U5EUC3 A0A195B7P9 A0A336MX53 W8CE95 A0A1I8NWH0 A0A023FWK3 F4WZL4 A0A131Y8E8 E1ZV52 A0A1B6GYD5 A0A1L8DVA3 A0A1S3DEK1 A0A0K8RQL2 B4J367 A0A0J7KPV6 A0A0M4E9J5 B4HV51 B4KZW8 T1DJV8 A0A1W4UGS4 B4PK62 A0A1B6JY16 A0A1B6GPZ7 B4LFC2 A0A1Z5L733 B4QKC9 A0A293M4P5 Q17BN4 A0A1A9X2U7 B5M0L7 B3NFE9 B4MLE4 A0A023EHB4 A0A182GLI4 A0A182PIN3 A0A1A9V594 A0A1E1WWR6
A0A212EYZ8 A0A2H1WR08 A0A2J7QF43 A0A0A9Z8G3 A0A0T6BG91 A0A1B6D4C1 A0A2P8XF57 A0A0N0BKM3 A0A2A3E7A7 V9IGH9 A0A088ATP6 V5IAW8 A0A2R7VWN8 A0A0L7L6S7 A0A310SHZ7 R4WDL8 D6W9A1 A0A1B6GK49 A0A158NGR7 A0A151XAC3 A0A195DXV4 A0A0L7QLH6 A0A026VX70 A0A1W4WEQ8 A0A151IFL0 A0A224XZ92 E9ILD6 J3JX07 K7IYD2 R4G882 A0A232F8L8 A0A0P4VSK9 A6YPS1 E2B3V1 A0A1B6M517 A0A1Y1MPL5 A0A131XD43 A0A1B0GLN9 A0A1Y1MHT7 A0A0K8W5Y6 A0A0K8TTI0 A0A224Z157 A0A131YW60 L7M309 A0A0L0CCD8 A0A1B0D2K8 A0A154P9T5 A0A0A1XFG0 N6TUS2 A0A034WJ33 A0A195FFC6 A0A023GF27 A0A023FFJ2 A0A087TGC9 T1P8V6 G3MLV9 A0A1L8ECA1 U5EUC3 A0A195B7P9 A0A336MX53 W8CE95 A0A1I8NWH0 A0A023FWK3 F4WZL4 A0A131Y8E8 E1ZV52 A0A1B6GYD5 A0A1L8DVA3 A0A1S3DEK1 A0A0K8RQL2 B4J367 A0A0J7KPV6 A0A0M4E9J5 B4HV51 B4KZW8 T1DJV8 A0A1W4UGS4 B4PK62 A0A1B6JY16 A0A1B6GPZ7 B4LFC2 A0A1Z5L733 B4QKC9 A0A293M4P5 Q17BN4 A0A1A9X2U7 B5M0L7 B3NFE9 B4MLE4 A0A023EHB4 A0A182GLI4 A0A182PIN3 A0A1A9V594 A0A1E1WWR6
PDB
5M73
E-value=4.17871e-29,
Score=313
Ontologies
GO
Topology
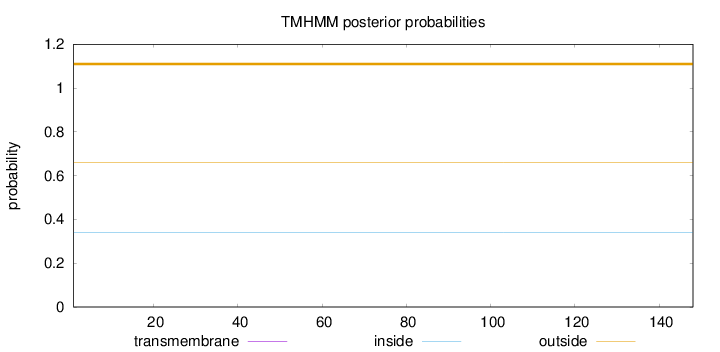
Length:
148
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00028
Exp number, first 60 AAs:
0.00028
Total prob of N-in:
0.33863
outside
1 - 148
Population Genetic Test Statistics
Pi
195.833499
Theta
162.94104
Tajima's D
0.651737
CLR
0.000459
CSRT
0.554522273886306
Interpretation
Uncertain
