Gene
KWMTBOMO05029
Pre Gene Modal
BGIBMGA012529
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_toll-like_receptor_8_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.784
Sequence
CDS
ATGGATATTATCACACGACTTTCCCGCTTCAAATATGGCGCCTACGTCCTTTTGGTCATCATCCATATTCCGATACTGCAAGGAGAAGAACAATCGCAATGTCCAAGTCTGTCAGAAAATCCGATTTGTCCTTGTTACAATTTTAAGGAAGGTCTTTTTCTGGAATGTCCGAGTGCCTCTCCTGGTGTTGTGAAAAATGTGTTATCAAAAATAAAAGGAACAATACAATCTTTATCGATTTACGATTTGGAGAGTTCGACCACTGAACTGAAAGCAGAACTTTTCCCGCCTAAAATAAAAATTTCGAATTTACAAATCTCCCAATCAAACATAGATAAAGGTAATCGTTTAACTACAATTGGCGATGTATTCCCAAACAGAAATTCATCTTTAGTTTCAATTCAGCTCGGGGCGAATTCTTTATCATTGTTGAAAAATAGCTCTCTTCGAGGACAAGTTAACGTTCAAATAATGTGGCTGAGCCAAAATAATATCAATATGTTAACTTCGAATCTTTTTGATGATTTGTTAAATATACAGAGGCTGTACCTTAAAAATAATAGTATTATACATATAGAAAGGGCATTTTCAAAACTGCACAACTTGGAGTTCCTCGATTTGAGTCAAAATAAATTAGAAAATGTCCCTCGAGAAAGGCTTCAGGGGTTATATTCAACACGGCTACTAAACATCTCCCAGAACATTATAAGGGAGCTAGAAGAATTCACCGCAGATTTGCAAAGCCTACAAAAAATTGACTTATCCTATAACCACATTACAAGAATTCATAGAGAAATATTTCATCGGCTACATAATCTTGTGGAGTTGAACTTAAGTGGAAATTGGCTATCAGCTATTTCTACAGATGCTTTTAAACATATGCACAAAATATCTGTTATAGACTTGAGTCATAATTACTTCGAAACAATTCAAATAAAAATGTTAACTTCATTGGAGTCTCAGATTAAAGCATTATCATTAGATGAGAATCCTTTCACATGCAATTGTGAATCACAGGATGTCTGGCGGTGGATGCAAGATCACTATAAAATTATTATTAAAGGAAGCTACAATTTGCGATGTGAACATCCAGAAGATATACACGGGTATAGGTTCCTCGAGCTTCCACATCAGAAATTATGCGATGTACCAATTGTAATTCGTATTGCAATTCAAGACATTCAAACATATTCTGTTGTCGTTTCTTGGCAAAGTCGAAATCAAACCGGCTTGAACGGATTTCAAGTGGCTTATTTCCCCGAGGAGAATCCTGGCGCTATACGAGGTAAGACGATAGGTGCCGGAATGAGATCTACAAGGTTAAATCATTTAACGCCAGGATCAAGGTACATTATTTGCGTATTAGCTTACGGGAATTTCAGTCCTACAACATCATTAGACTCCTCGGTCCCAGATGCCAGGGTAGCGCCGTCACCCGCTAACAAAAACAGCATATACGGAGATGAACAAATAATGAATCATCTAAAATCTTATATGAATGATTCCATGACTAGTAAGTGTACAAGTGTTAGCACTATTGAAATATTCGGTGCAGCTATTGACAGCCCATTTTCTAATACATATATGGGCATCGCTGATATTCTGACCAGGAGACTCAGTTTAGTAGTCGGTTGCTGCATTGGATTTATCGTGTTCGTCGTTTTGGTATCAGCATTAGGTTACATCAAAAGTAAAAAGCGTCAAGTGATTGCGAAAAGTGAATCTCAACAGCCCCCACAATATATTTCGTATGACAACTTCAATACTCCAAGTGTAGAAGTTCAAACAACAGATATGGATATAAATACTATTACGGACAAAACGAAGTCTCAATGCAAGCATGAGTGA
Protein
MDIITRLSRFKYGAYVLLVIIHIPILQGEEQSQCPSLSENPICPCYNFKEGLFLECPSASPGVVKNVLSKIKGTIQSLSIYDLESSTTELKAELFPPKIKISNLQISQSNIDKGNRLTTIGDVFPNRNSSLVSIQLGANSLSLLKNSSLRGQVNVQIMWLSQNNINMLTSNLFDDLLNIQRLYLKNNSIIHIERAFSKLHNLEFLDLSQNKLENVPRERLQGLYSTRLLNISQNIIRELEEFTADLQSLQKIDLSYNHITRIHREIFHRLHNLVELNLSGNWLSAISTDAFKHMHKISVIDLSHNYFETIQIKMLTSLESQIKALSLDENPFTCNCESQDVWRWMQDHYKIIIKGSYNLRCEHPEDIHGYRFLELPHQKLCDVPIVIRIAIQDIQTYSVVVSWQSRNQTGLNGFQVAYFPEENPGAIRGKTIGAGMRSTRLNHLTPGSRYIICVLAYGNFSPTTSLDSSVPDARVAPSPANKNSIYGDEQIMNHLKSYMNDSMTSKCTSVSTIEIFGAAIDSPFSNTYMGIADILTRRLSLVVGCCIGFIVFVVLVSALGYIKSKKRQVIAKSESQQPPQYISYDNFNTPSVEVQTTDMDINTITDKTKSQCKHE
Summary
Uniprot
A0A2A4IYZ0
A0A2H1WXC0
A0A3S2N878
A0A0L7KML4
A0A0N1PFN8
A0A1W4X7P6
+ More
A0A2J7Q2A1 A0A139WMU4 D6W995 A0A067QYH8 A0A151WEP6 A0A151IYZ8 A0A182V7C2 F4X3A9 A0A158NEJ6 K7J4W3 A0A2P8XAD9 A0A026WIB3 A0A3L8E027 A0A182SN70 A0A195FI09 A0A0L7RHL2 A0A182HEB3 A0A2H8TP25 K7J4W0 E2APU1 U4UII3 A0A182IAZ4 A0A182W658 E2BAJ0 A0A182MPR5 A0A182PBH4 A0A182Q339 F4WEC8 A0A151WEM2 A0A151J9Q9 A0A195BDK3 A0A195BEP1 A0A158NDY6 B0WH32 A0A088AS96 A0A026X402 A0A0J7P243 A0A0N1ITQ9 A0A182YF08 E9IHM6 A0A0C9RCR5 A0A182R282 T1HEM2 A0A195C217 A0A0J7KRN9 A0A154P712 A0A084VFT8 A0A1B0D2X1 A0A195C1E2 A0A2A3ETJ9 A0A088AS51 A0A0C9PHN7 A0A182N0Q5 E2APT3 A0A182JMC3 A0A195FGY5 J9K8S1 A0A1J1IZZ4 A0A2S2QRI2 A0A182FIM6 A0A2S2P545 W5JSP3 Q059E0 A0A1A9WPL0 B4PZM2 A0A0N0BHE4 A0A1A9UUM2 B4R452 B4IKP1
A0A2J7Q2A1 A0A139WMU4 D6W995 A0A067QYH8 A0A151WEP6 A0A151IYZ8 A0A182V7C2 F4X3A9 A0A158NEJ6 K7J4W3 A0A2P8XAD9 A0A026WIB3 A0A3L8E027 A0A182SN70 A0A195FI09 A0A0L7RHL2 A0A182HEB3 A0A2H8TP25 K7J4W0 E2APU1 U4UII3 A0A182IAZ4 A0A182W658 E2BAJ0 A0A182MPR5 A0A182PBH4 A0A182Q339 F4WEC8 A0A151WEM2 A0A151J9Q9 A0A195BDK3 A0A195BEP1 A0A158NDY6 B0WH32 A0A088AS96 A0A026X402 A0A0J7P243 A0A0N1ITQ9 A0A182YF08 E9IHM6 A0A0C9RCR5 A0A182R282 T1HEM2 A0A195C217 A0A0J7KRN9 A0A154P712 A0A084VFT8 A0A1B0D2X1 A0A195C1E2 A0A2A3ETJ9 A0A088AS51 A0A0C9PHN7 A0A182N0Q5 E2APT3 A0A182JMC3 A0A195FGY5 J9K8S1 A0A1J1IZZ4 A0A2S2QRI2 A0A182FIM6 A0A2S2P545 W5JSP3 Q059E0 A0A1A9WPL0 B4PZM2 A0A0N0BHE4 A0A1A9UUM2 B4R452 B4IKP1
Pubmed
EMBL
NWSH01004405
PCG65157.1
ODYU01011706
SOQ57616.1
RSAL01000217
RVE44103.1
+ More
JTDY01008523 KOB64533.1 KQ461133 KPJ08895.1 NEVH01019373 PNF22712.1 KQ971312 KYB29176.1 EEZ99221.1 KK852874 KDR14538.1 KQ983238 KYQ46296.1 KQ980731 KYN13972.1 GL888609 EGI59090.1 ADTU01000243 ADTU01000244 AAZX01000454 PYGN01020773 PSN28966.1 KK107193 EZA55703.1 QOIP01000002 RLU26071.1 KQ981606 KYN39624.1 KQ414590 KOC70309.1 JXUM01131322 JXUM01131323 KQ567697 KXJ69314.1 GFXV01003276 GFXV01004122 GFXV01007411 GFXV01007491 MBW15081.1 MBW15927.1 MBW19216.1 MBW19296.1 AAZX01008445 GL441581 EFN64567.1 KB632314 ERL92193.1 APCN01001389 GL446759 EFN87284.1 AXCM01003610 AXCN02001398 GL888102 EGI67566.1 KYQ46304.1 KQ979388 KYN21757.1 KQ976509 KYM82638.1 KYM82630.1 ADTU01000178 DS231932 EDS27483.1 KK107017 EZA62828.1 RLU25539.1 LBMM01000339 KMR00321.1 KQ435753 KOX76128.1 GL763289 EFZ19953.1 GBYB01010917 GBYB01010918 JAG80684.1 JAG80685.1 ACPB03020071 KQ978350 KYM94655.1 LBMM01004009 KMQ92904.1 KQ434829 KZC07719.1 ATLV01012462 KE524793 KFB36832.1 AJVK01002906 AJVK01002907 KYM94662.1 KZ288186 PBC34824.1 GBYB01000383 JAG70150.1 EFN64559.1 KYN39631.1 ABLF02027459 ABLF02027460 CVRI01000063 CRL04742.1 GGMS01010937 MBY80140.1 GGMR01012002 MBY24621.1 ADMH02000273 ETN67161.1 BT029028 ABJ16961.1 CM000162 EDX03150.2 KRK07151.1 KRK07152.1 KOX76118.1 CM000366 EDX18574.1 CH480854 EDW51655.1
JTDY01008523 KOB64533.1 KQ461133 KPJ08895.1 NEVH01019373 PNF22712.1 KQ971312 KYB29176.1 EEZ99221.1 KK852874 KDR14538.1 KQ983238 KYQ46296.1 KQ980731 KYN13972.1 GL888609 EGI59090.1 ADTU01000243 ADTU01000244 AAZX01000454 PYGN01020773 PSN28966.1 KK107193 EZA55703.1 QOIP01000002 RLU26071.1 KQ981606 KYN39624.1 KQ414590 KOC70309.1 JXUM01131322 JXUM01131323 KQ567697 KXJ69314.1 GFXV01003276 GFXV01004122 GFXV01007411 GFXV01007491 MBW15081.1 MBW15927.1 MBW19216.1 MBW19296.1 AAZX01008445 GL441581 EFN64567.1 KB632314 ERL92193.1 APCN01001389 GL446759 EFN87284.1 AXCM01003610 AXCN02001398 GL888102 EGI67566.1 KYQ46304.1 KQ979388 KYN21757.1 KQ976509 KYM82638.1 KYM82630.1 ADTU01000178 DS231932 EDS27483.1 KK107017 EZA62828.1 RLU25539.1 LBMM01000339 KMR00321.1 KQ435753 KOX76128.1 GL763289 EFZ19953.1 GBYB01010917 GBYB01010918 JAG80684.1 JAG80685.1 ACPB03020071 KQ978350 KYM94655.1 LBMM01004009 KMQ92904.1 KQ434829 KZC07719.1 ATLV01012462 KE524793 KFB36832.1 AJVK01002906 AJVK01002907 KYM94662.1 KZ288186 PBC34824.1 GBYB01000383 JAG70150.1 EFN64559.1 KYN39631.1 ABLF02027459 ABLF02027460 CVRI01000063 CRL04742.1 GGMS01010937 MBY80140.1 GGMR01012002 MBY24621.1 ADMH02000273 ETN67161.1 BT029028 ABJ16961.1 CM000162 EDX03150.2 KRK07151.1 KRK07152.1 KOX76118.1 CM000366 EDX18574.1 CH480854 EDW51655.1
Proteomes
UP000218220
UP000283053
UP000037510
UP000053240
UP000192223
UP000235965
+ More
UP000007266 UP000027135 UP000075809 UP000078492 UP000075903 UP000007755 UP000005205 UP000002358 UP000245037 UP000053097 UP000279307 UP000075901 UP000078541 UP000053825 UP000069940 UP000249989 UP000000311 UP000030742 UP000075840 UP000075920 UP000008237 UP000075883 UP000075885 UP000075886 UP000078540 UP000002320 UP000005203 UP000036403 UP000053105 UP000076408 UP000075900 UP000015103 UP000078542 UP000076502 UP000030765 UP000092462 UP000242457 UP000075884 UP000075880 UP000007819 UP000183832 UP000069272 UP000000673 UP000091820 UP000002282 UP000078200 UP000000304 UP000001292
UP000007266 UP000027135 UP000075809 UP000078492 UP000075903 UP000007755 UP000005205 UP000002358 UP000245037 UP000053097 UP000279307 UP000075901 UP000078541 UP000053825 UP000069940 UP000249989 UP000000311 UP000030742 UP000075840 UP000075920 UP000008237 UP000075883 UP000075885 UP000075886 UP000078540 UP000002320 UP000005203 UP000036403 UP000053105 UP000076408 UP000075900 UP000015103 UP000078542 UP000076502 UP000030765 UP000092462 UP000242457 UP000075884 UP000075880 UP000007819 UP000183832 UP000069272 UP000000673 UP000091820 UP000002282 UP000078200 UP000000304 UP000001292
PRIDE
Interpro
SUPFAM
SSF49265
SSF49265
Gene 3D
CDD
ProteinModelPortal
A0A2A4IYZ0
A0A2H1WXC0
A0A3S2N878
A0A0L7KML4
A0A0N1PFN8
A0A1W4X7P6
+ More
A0A2J7Q2A1 A0A139WMU4 D6W995 A0A067QYH8 A0A151WEP6 A0A151IYZ8 A0A182V7C2 F4X3A9 A0A158NEJ6 K7J4W3 A0A2P8XAD9 A0A026WIB3 A0A3L8E027 A0A182SN70 A0A195FI09 A0A0L7RHL2 A0A182HEB3 A0A2H8TP25 K7J4W0 E2APU1 U4UII3 A0A182IAZ4 A0A182W658 E2BAJ0 A0A182MPR5 A0A182PBH4 A0A182Q339 F4WEC8 A0A151WEM2 A0A151J9Q9 A0A195BDK3 A0A195BEP1 A0A158NDY6 B0WH32 A0A088AS96 A0A026X402 A0A0J7P243 A0A0N1ITQ9 A0A182YF08 E9IHM6 A0A0C9RCR5 A0A182R282 T1HEM2 A0A195C217 A0A0J7KRN9 A0A154P712 A0A084VFT8 A0A1B0D2X1 A0A195C1E2 A0A2A3ETJ9 A0A088AS51 A0A0C9PHN7 A0A182N0Q5 E2APT3 A0A182JMC3 A0A195FGY5 J9K8S1 A0A1J1IZZ4 A0A2S2QRI2 A0A182FIM6 A0A2S2P545 W5JSP3 Q059E0 A0A1A9WPL0 B4PZM2 A0A0N0BHE4 A0A1A9UUM2 B4R452 B4IKP1
A0A2J7Q2A1 A0A139WMU4 D6W995 A0A067QYH8 A0A151WEP6 A0A151IYZ8 A0A182V7C2 F4X3A9 A0A158NEJ6 K7J4W3 A0A2P8XAD9 A0A026WIB3 A0A3L8E027 A0A182SN70 A0A195FI09 A0A0L7RHL2 A0A182HEB3 A0A2H8TP25 K7J4W0 E2APU1 U4UII3 A0A182IAZ4 A0A182W658 E2BAJ0 A0A182MPR5 A0A182PBH4 A0A182Q339 F4WEC8 A0A151WEM2 A0A151J9Q9 A0A195BDK3 A0A195BEP1 A0A158NDY6 B0WH32 A0A088AS96 A0A026X402 A0A0J7P243 A0A0N1ITQ9 A0A182YF08 E9IHM6 A0A0C9RCR5 A0A182R282 T1HEM2 A0A195C217 A0A0J7KRN9 A0A154P712 A0A084VFT8 A0A1B0D2X1 A0A195C1E2 A0A2A3ETJ9 A0A088AS51 A0A0C9PHN7 A0A182N0Q5 E2APT3 A0A182JMC3 A0A195FGY5 J9K8S1 A0A1J1IZZ4 A0A2S2QRI2 A0A182FIM6 A0A2S2P545 W5JSP3 Q059E0 A0A1A9WPL0 B4PZM2 A0A0N0BHE4 A0A1A9UUM2 B4R452 B4IKP1
PDB
4OQT
E-value=3.57912e-16,
Score=209
Ontologies
GO
Topology
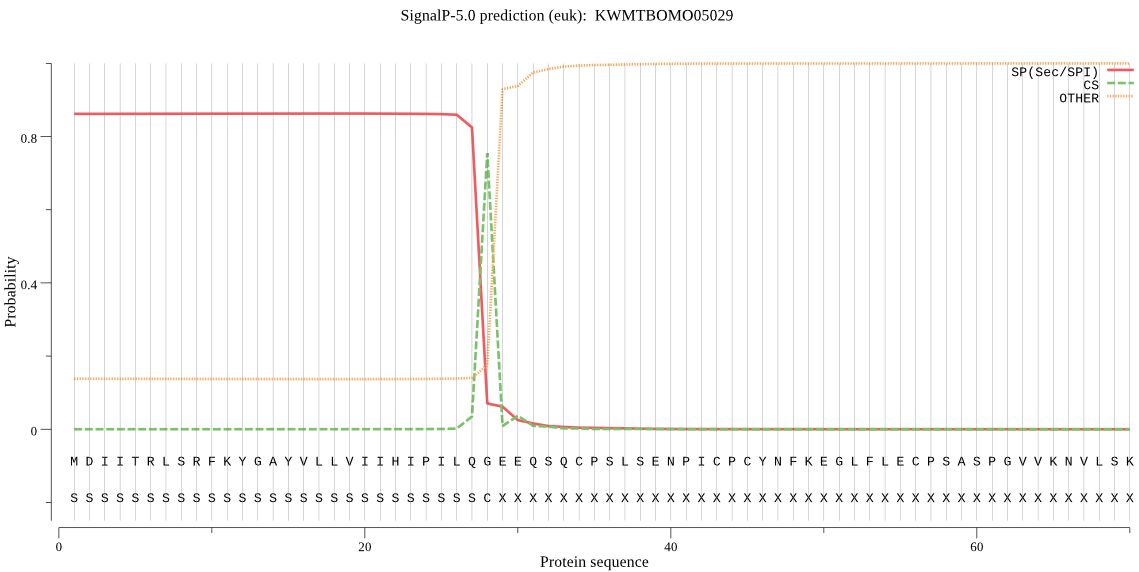
SignalP
Position: 1 - 28,
Likelihood: 0.863677
Length:
615
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.96293
Exp number, first 60 AAs:
5.02905
Total prob of N-in:
0.27167
outside
1 - 539
TMhelix
540 - 562
inside
563 - 615

Population Genetic Test Statistics
Pi
218.283889
Theta
182.467068
Tajima's D
0.70238
CLR
0.270717
CSRT
0.577571121443928
Interpretation
Uncertain
