Gene
KWMTBOMO05024
Pre Gene Modal
BGIBMGA012527
Annotation
putative_RDH13_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 1.179 PlasmaMembrane Reliability : 1.619
Sequence
CDS
ATGTTTTGGTGGATGGTATTAGCCGTCATTGGAGTTATAGTGCTATTAGTAAAGGTGCATCAAAAACTGACGCTTGGAATATGTAAGTCGAGTGCGCATATGGTGGGTAAAGTAGTTATAGTGACTGGAGGAAACAGTGGCATTGGTTTTGAGACAGCAAAAGACTTGGCTTGTAGAGGTGCACGAGTTATACTAGCTTGTAGAAGTGTCCGCAAAGCTGAAATAGTTAAAGAAGAATTGATTAAAGCTAGTGGTAACACAGATATTATTTGTAAGCAGTTAGATCTTTCATCGCTCGAGTCTGTTAGAAATTTTTGCGAAGATATAGATAAAAACGAAAAACGTTTGGACGTTCTTATTAACAATGCGGGTGCCGGTGGATTAGGCAATTACAAAACGGCGGACGGATTACATGTTGGAATGCAAGTTAATTATTTTGCTCCGTTTTTATTAACGTGCCTTCTGTTGCCACTTCTAAGAAGATCCGAGCCGAGTCGAATTATTAATGTGTCCTCTATGTTGCACAAATACGGAGAACTGGATTTTGAAAATTTGAATATGGAAAAATACTGGAGTGACTATTTAGTATATGCAAATAGTAAATTGTTCTTGAACTTAATGACTTTAGAATTGAGTCGTAGATTGAATGGGTCTAGGGTTACTGTGAACTGCTTACATCCGGGTATAGCTGCTACTAACTTAATGAGAAACGTGCCCAGTGAAATTTTGAAGAAGATAATTTCGAAATCAATAGGCTTTATGTTCCAGAGTGCATGGGAAGCGTCTCAAACTTCTATCTATTTGTCAGTAGCACCAGAAATTCAAGATATTAGTGGAAAATATTTCAGTCATTGCTGTGAAACAAAGCCTTCGAAGCTATCTCTCGATGTTGAAAAAGCAAAAAAGCTTTGGGACGAATCAGAAAAATTAGTGAAGTTCACGATATCAGAAAATTAG
Protein
MFWWMVLAVIGVIVLLVKVHQKLTLGICKSSAHMVGKVVIVTGGNSGIGFETAKDLACRGARVILACRSVRKAEIVKEELIKASGNTDIICKQLDLSSLESVRNFCEDIDKNEKRLDVLINNAGAGGLGNYKTADGLHVGMQVNYFAPFLLTCLLLPLLRRSEPSRIINVSSMLHKYGELDFENLNMEKYWSDYLVYANSKLFLNLMTLELSRRLNGSRVTVNCLHPGIAATNLMRNVPSEILKKIISKSIGFMFQSAWEASQTSIYLSVAPEIQDISGKYFSHCCETKPSKLSLDVEKAKKLWDESEKLVKFTISEN
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JSL6
A0A2H1W7C7
A0A2A4ITG8
A0A0L7KVJ8
A0A0N0PAN9
A0A0N0PAY1
+ More
A0A212F5B7 A0A0N1IF98 A0A0N0PAB9 A0A0L7KV18 A0A2A4IXN8 A0A2H1W6X7 A0A2J7PGY1 I4DQA1 A0A1Q3G5H8 A0A0N1ICN4 A0A0N0PAY2 A0A0N0PB70 A0A2J7PGT0 H9JSR3 A0A0N0PAC1 A0A0N8E5H6 A0A1B6DPI9 A0A2J7PGT7 A0A1Q3G5G7 A0A067QUP0 R4FLL5 A0A1B6FRX7 A0A0P5DLU5 A0A151IL06 H9GAK1 A0A069DST5 A0A151WEM9 A0A195FH86 E2AM66 A0A2J7PGU9 D6X0B3 A0A151IVL0 A0A0K8URJ7 A0A0P6BK02 A0A195BEN0 A0A158NKK8 A0A1W4XHF8 W8BZ25 A0A034W5L3 A0A1B6IV52 A0A0L7RHT2 A0A0N1IDL3 K7IP74 E9GMH1 A0A1B6LG81 E2BNF8 A0A0P5JEC0 A0A0A1XDP5 V9KU50 A0A224XFL4 Q17GA4 A0A0P5JH40 V4AKA2 K1R790 A0A2I4AU28 A0A026WH33 A0A3Q2WGP4 K1Q9G0 A0A3Q4G365 A0A087TBA3 A0A3P8PS94 A0A3P9BNR6 A0A2I4AU46 A0A2C9K9F2 W8BRX4 A0A0K8VZD3 B0XLA9 A0A034W3X1 A7RNX1 A0A0P5LS57 A0A1W4WGA8 R4GAG6 A0A0S7KYR3 A0A182VIF5 A0A182LH67 A0A182HY29 E9GHT3 A0A0A1X7Y5 A0A2P6L863 B4M3Z8 A0A1W4WGA3 A0A182U7J4 W4ZIF1 A0A3B4GRP8 I3J2X8 A0A2J7PGU3 A0A0L7L5N4 A0A2J7QDU2 A0A0J7L004 A0A0P5UI40 A0A3Q1EP65 A0A023F7P3 A0A3Q2TZ83 F4X379
A0A212F5B7 A0A0N1IF98 A0A0N0PAB9 A0A0L7KV18 A0A2A4IXN8 A0A2H1W6X7 A0A2J7PGY1 I4DQA1 A0A1Q3G5H8 A0A0N1ICN4 A0A0N0PAY2 A0A0N0PB70 A0A2J7PGT0 H9JSR3 A0A0N0PAC1 A0A0N8E5H6 A0A1B6DPI9 A0A2J7PGT7 A0A1Q3G5G7 A0A067QUP0 R4FLL5 A0A1B6FRX7 A0A0P5DLU5 A0A151IL06 H9GAK1 A0A069DST5 A0A151WEM9 A0A195FH86 E2AM66 A0A2J7PGU9 D6X0B3 A0A151IVL0 A0A0K8URJ7 A0A0P6BK02 A0A195BEN0 A0A158NKK8 A0A1W4XHF8 W8BZ25 A0A034W5L3 A0A1B6IV52 A0A0L7RHT2 A0A0N1IDL3 K7IP74 E9GMH1 A0A1B6LG81 E2BNF8 A0A0P5JEC0 A0A0A1XDP5 V9KU50 A0A224XFL4 Q17GA4 A0A0P5JH40 V4AKA2 K1R790 A0A2I4AU28 A0A026WH33 A0A3Q2WGP4 K1Q9G0 A0A3Q4G365 A0A087TBA3 A0A3P8PS94 A0A3P9BNR6 A0A2I4AU46 A0A2C9K9F2 W8BRX4 A0A0K8VZD3 B0XLA9 A0A034W3X1 A7RNX1 A0A0P5LS57 A0A1W4WGA8 R4GAG6 A0A0S7KYR3 A0A182VIF5 A0A182LH67 A0A182HY29 E9GHT3 A0A0A1X7Y5 A0A2P6L863 B4M3Z8 A0A1W4WGA3 A0A182U7J4 W4ZIF1 A0A3B4GRP8 I3J2X8 A0A2J7PGU3 A0A0L7L5N4 A0A2J7QDU2 A0A0J7L004 A0A0P5UI40 A0A3Q1EP65 A0A023F7P3 A0A3Q2TZ83 F4X379
Pubmed
EMBL
BABH01032240
ODYU01006737
SOQ48856.1
NWSH01008708
PCG62440.1
JTDY01005329
+ More
KOB67106.1 KQ458665 KPJ05596.1 KQ461133 KPJ08898.1 AGBW02010210 OWR48927.1 KPJ08897.1 KPJ05595.1 KOB67107.1 NWSH01005060 PCG64471.1 SOQ48855.1 NEVH01025161 PNF15592.1 AK404009 BAM20091.1 GEYN01000183 JAV44806.1 KPJ05597.1 KPJ08901.1 KPJ08899.1 PNF15543.1 BABH01032245 KPJ05598.1 GDIQ01059860 JAN34877.1 GEDC01009721 JAS27577.1 PNF15542.1 GEYN01000196 JAV44793.1 KK853387 KDR07942.1 GAHY01001333 JAA76177.1 GECZ01025002 GECZ01017006 JAS44767.1 JAS52763.1 GDIP01157800 JAJ65602.1 KQ977151 KYN05278.1 AAWZ02022819 GBGD01002103 JAC86786.1 KQ983238 KYQ46262.1 KQ981606 KYN39592.1 GL440715 EFN65472.1 PNF15560.1 KQ971372 EFA10520.1 KQ980895 KYN11766.1 GDHF01023108 JAI29206.1 GDIP01013477 LRGB01000243 JAM90238.1 KZS20137.1 KQ976509 KYM82669.1 ADTU01000291 GAMC01004622 JAC01934.1 GAKP01009537 JAC49415.1 GECU01016890 JAS90816.1 KQ414590 KOC70369.1 KPJ08900.1 GL732552 EFX79364.1 GEBQ01017270 JAT22707.1 GL449414 EFN82737.1 GDIQ01201309 JAK50416.1 GBXI01016837 GBXI01005231 JAC97454.1 JAD09061.1 JW869425 AFP01943.1 GFTR01005159 JAW11267.1 CH477264 EAT45601.1 EAT45602.1 GDIQ01200010 JAK51715.1 KB201701 ESO95165.1 JH818186 EKC41628.1 KK107250 QOIP01000002 EZA54384.1 RLU25994.1 JH817020 EKC30553.1 KK114425 KFM62392.1 GAMC01004623 JAC01933.1 GDHF01008354 JAI43960.1 DS234386 EDS33797.1 GAKP01009538 JAC49414.1 DS469524 EDO46815.1 GDIQ01166025 JAK85700.1 GBYX01192259 JAO82412.1 APCN01004485 GL732545 EFX80894.1 GBXI01006878 JAD07414.1 MWRG01001082 PRD34763.1 CH940652 EDW59359.1 AAGJ04078790 AERX01000040 PNF15541.1 JTDY01002853 KOB70599.1 NEVH01015814 PNF26749.1 LBMM01001639 KMQ95961.1 GDIP01112562 JAL91152.1 GBBI01001321 JAC17391.1 GL888609 EGI59060.1
KOB67106.1 KQ458665 KPJ05596.1 KQ461133 KPJ08898.1 AGBW02010210 OWR48927.1 KPJ08897.1 KPJ05595.1 KOB67107.1 NWSH01005060 PCG64471.1 SOQ48855.1 NEVH01025161 PNF15592.1 AK404009 BAM20091.1 GEYN01000183 JAV44806.1 KPJ05597.1 KPJ08901.1 KPJ08899.1 PNF15543.1 BABH01032245 KPJ05598.1 GDIQ01059860 JAN34877.1 GEDC01009721 JAS27577.1 PNF15542.1 GEYN01000196 JAV44793.1 KK853387 KDR07942.1 GAHY01001333 JAA76177.1 GECZ01025002 GECZ01017006 JAS44767.1 JAS52763.1 GDIP01157800 JAJ65602.1 KQ977151 KYN05278.1 AAWZ02022819 GBGD01002103 JAC86786.1 KQ983238 KYQ46262.1 KQ981606 KYN39592.1 GL440715 EFN65472.1 PNF15560.1 KQ971372 EFA10520.1 KQ980895 KYN11766.1 GDHF01023108 JAI29206.1 GDIP01013477 LRGB01000243 JAM90238.1 KZS20137.1 KQ976509 KYM82669.1 ADTU01000291 GAMC01004622 JAC01934.1 GAKP01009537 JAC49415.1 GECU01016890 JAS90816.1 KQ414590 KOC70369.1 KPJ08900.1 GL732552 EFX79364.1 GEBQ01017270 JAT22707.1 GL449414 EFN82737.1 GDIQ01201309 JAK50416.1 GBXI01016837 GBXI01005231 JAC97454.1 JAD09061.1 JW869425 AFP01943.1 GFTR01005159 JAW11267.1 CH477264 EAT45601.1 EAT45602.1 GDIQ01200010 JAK51715.1 KB201701 ESO95165.1 JH818186 EKC41628.1 KK107250 QOIP01000002 EZA54384.1 RLU25994.1 JH817020 EKC30553.1 KK114425 KFM62392.1 GAMC01004623 JAC01933.1 GDHF01008354 JAI43960.1 DS234386 EDS33797.1 GAKP01009538 JAC49414.1 DS469524 EDO46815.1 GDIQ01166025 JAK85700.1 GBYX01192259 JAO82412.1 APCN01004485 GL732545 EFX80894.1 GBXI01006878 JAD07414.1 MWRG01001082 PRD34763.1 CH940652 EDW59359.1 AAGJ04078790 AERX01000040 PNF15541.1 JTDY01002853 KOB70599.1 NEVH01015814 PNF26749.1 LBMM01001639 KMQ95961.1 GDIP01112562 JAL91152.1 GBBI01001321 JAC17391.1 GL888609 EGI59060.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000235965 UP000027135 UP000078542 UP000001646 UP000075809 UP000078541 UP000000311 UP000007266 UP000078492 UP000076858 UP000078540 UP000005205 UP000192223 UP000053825 UP000002358 UP000000305 UP000008237 UP000008820 UP000030746 UP000005408 UP000192220 UP000053097 UP000279307 UP000264840 UP000261580 UP000054359 UP000265100 UP000265160 UP000076420 UP000002320 UP000001593 UP000075903 UP000075882 UP000075840 UP000008792 UP000075902 UP000007110 UP000261460 UP000005207 UP000036403 UP000257200 UP000265000 UP000007755
UP000235965 UP000027135 UP000078542 UP000001646 UP000075809 UP000078541 UP000000311 UP000007266 UP000078492 UP000076858 UP000078540 UP000005205 UP000192223 UP000053825 UP000002358 UP000000305 UP000008237 UP000008820 UP000030746 UP000005408 UP000192220 UP000053097 UP000279307 UP000264840 UP000261580 UP000054359 UP000265100 UP000265160 UP000076420 UP000002320 UP000001593 UP000075903 UP000075882 UP000075840 UP000008792 UP000075902 UP000007110 UP000261460 UP000005207 UP000036403 UP000257200 UP000265000 UP000007755
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JSL6
A0A2H1W7C7
A0A2A4ITG8
A0A0L7KVJ8
A0A0N0PAN9
A0A0N0PAY1
+ More
A0A212F5B7 A0A0N1IF98 A0A0N0PAB9 A0A0L7KV18 A0A2A4IXN8 A0A2H1W6X7 A0A2J7PGY1 I4DQA1 A0A1Q3G5H8 A0A0N1ICN4 A0A0N0PAY2 A0A0N0PB70 A0A2J7PGT0 H9JSR3 A0A0N0PAC1 A0A0N8E5H6 A0A1B6DPI9 A0A2J7PGT7 A0A1Q3G5G7 A0A067QUP0 R4FLL5 A0A1B6FRX7 A0A0P5DLU5 A0A151IL06 H9GAK1 A0A069DST5 A0A151WEM9 A0A195FH86 E2AM66 A0A2J7PGU9 D6X0B3 A0A151IVL0 A0A0K8URJ7 A0A0P6BK02 A0A195BEN0 A0A158NKK8 A0A1W4XHF8 W8BZ25 A0A034W5L3 A0A1B6IV52 A0A0L7RHT2 A0A0N1IDL3 K7IP74 E9GMH1 A0A1B6LG81 E2BNF8 A0A0P5JEC0 A0A0A1XDP5 V9KU50 A0A224XFL4 Q17GA4 A0A0P5JH40 V4AKA2 K1R790 A0A2I4AU28 A0A026WH33 A0A3Q2WGP4 K1Q9G0 A0A3Q4G365 A0A087TBA3 A0A3P8PS94 A0A3P9BNR6 A0A2I4AU46 A0A2C9K9F2 W8BRX4 A0A0K8VZD3 B0XLA9 A0A034W3X1 A7RNX1 A0A0P5LS57 A0A1W4WGA8 R4GAG6 A0A0S7KYR3 A0A182VIF5 A0A182LH67 A0A182HY29 E9GHT3 A0A0A1X7Y5 A0A2P6L863 B4M3Z8 A0A1W4WGA3 A0A182U7J4 W4ZIF1 A0A3B4GRP8 I3J2X8 A0A2J7PGU3 A0A0L7L5N4 A0A2J7QDU2 A0A0J7L004 A0A0P5UI40 A0A3Q1EP65 A0A023F7P3 A0A3Q2TZ83 F4X379
A0A212F5B7 A0A0N1IF98 A0A0N0PAB9 A0A0L7KV18 A0A2A4IXN8 A0A2H1W6X7 A0A2J7PGY1 I4DQA1 A0A1Q3G5H8 A0A0N1ICN4 A0A0N0PAY2 A0A0N0PB70 A0A2J7PGT0 H9JSR3 A0A0N0PAC1 A0A0N8E5H6 A0A1B6DPI9 A0A2J7PGT7 A0A1Q3G5G7 A0A067QUP0 R4FLL5 A0A1B6FRX7 A0A0P5DLU5 A0A151IL06 H9GAK1 A0A069DST5 A0A151WEM9 A0A195FH86 E2AM66 A0A2J7PGU9 D6X0B3 A0A151IVL0 A0A0K8URJ7 A0A0P6BK02 A0A195BEN0 A0A158NKK8 A0A1W4XHF8 W8BZ25 A0A034W5L3 A0A1B6IV52 A0A0L7RHT2 A0A0N1IDL3 K7IP74 E9GMH1 A0A1B6LG81 E2BNF8 A0A0P5JEC0 A0A0A1XDP5 V9KU50 A0A224XFL4 Q17GA4 A0A0P5JH40 V4AKA2 K1R790 A0A2I4AU28 A0A026WH33 A0A3Q2WGP4 K1Q9G0 A0A3Q4G365 A0A087TBA3 A0A3P8PS94 A0A3P9BNR6 A0A2I4AU46 A0A2C9K9F2 W8BRX4 A0A0K8VZD3 B0XLA9 A0A034W3X1 A7RNX1 A0A0P5LS57 A0A1W4WGA8 R4GAG6 A0A0S7KYR3 A0A182VIF5 A0A182LH67 A0A182HY29 E9GHT3 A0A0A1X7Y5 A0A2P6L863 B4M3Z8 A0A1W4WGA3 A0A182U7J4 W4ZIF1 A0A3B4GRP8 I3J2X8 A0A2J7PGU3 A0A0L7L5N4 A0A2J7QDU2 A0A0J7L004 A0A0P5UI40 A0A3Q1EP65 A0A023F7P3 A0A3Q2TZ83 F4X379
PDB
3RD5
E-value=7.31754e-22,
Score=255
Ontologies
GO
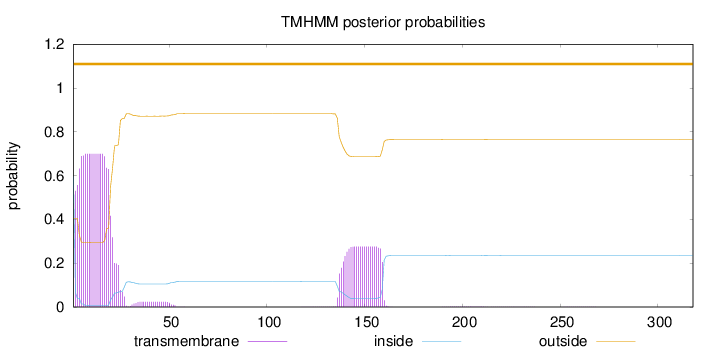
Topology
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
20.08164
Exp number, first 60 AAs:
14.04009
Total prob of N-in:
0.59578
POSSIBLE N-term signal
sequence
outside
1 - 318
Population Genetic Test Statistics
Pi
341.641043
Theta
185.614648
Tajima's D
2.66004
CLR
0.426541
CSRT
0.95925203739813
Interpretation
Uncertain
