Gene
KWMTBOMO05022
Pre Gene Modal
BGIBMGA012574
Annotation
PREDICTED:_retinol_dehydrogenase_11-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.309 Mitochondrial Reliability : 1.491
Sequence
CDS
ATGTGTAGGCATTTGGTCAGCAAAGTGGTGATAGTGACCGGAAGCAACAACGGTATAGGATACGAGACAGCAAAAGACTTAGCGGAGCGTGGAGCCCGGGTAATACTCGCTTGCAGGAACGATCTACGCGGTACCAAGGCCAGAGACCAGATCATTGCAGAATCAGGGAACCACGATGTGCATTATCGTCAGTTGGATTTAGCGTCATTTGGTTCTGTCAGAAGATTCGCTGAAGAATTCATTAAAACCGAAAATCGTTTAGACATTCTTATCAACAATGCTGGAGTACTCGTCGTGGACAATGTAAAGACGGAAAACGGCTTGTTTGTCGGTATGCAAGCAAATCACTTCGGGCCATTTCTGCTCACGAAGCTTCTTCTTCCGCTAATTAAATCGTCAGCTCCGAGTCGCATCATCAACGTTTCTTCCCTAGCCTACTCAAAAGGAATTATTGATTTTAACAATTTAAATTTGGAAAAAGAAACCCCGCAGTCATACAGTAAAATACAAGCATATAAAAACAGCAAGCTGTGTAATGTTCTAATGACCACTGAGTTGGCAAGACGGCTTGAAGGTACTAGAGTGACTGCTAATTGCTTACATCCCGGCTTAGTAAAAACTGAAATTGTAAATTCTGATAAACTGCCATTTTCGAAGTACTGGGTTCCTATCGCAAGTTTATTTGTTAAGACCCCTCGGGAAGGGGCTCAGACTTCAATATACCTGGCGGTGTCTCCTGAGGTAGAAAACGTAACCGGCTGCTATTTCGCCGATTGTCAGGTTAAAACTTTAACCGAAACGGGGCGTAATACTGAACTGGCTCGCCGTCTCTGGGAAGAGTCTGAAAAAATAATTGCGAAGAAATGA
Protein
MCRHLVSKVVIVTGSNNGIGYETAKDLAERGARVILACRNDLRGTKARDQIIAESGNHDVHYRQLDLASFGSVRRFAEEFIKTENRLDILINNAGVLVVDNVKTENGLFVGMQANHFGPFLLTKLLLPLIKSSAPSRIINVSSLAYSKGIIDFNNLNLEKETPQSYSKIQAYKNSKLCNVLMTTELARRLEGTRVTANCLHPGLVKTEIVNSDKLPFSKYWVPIASLFVKTPREGAQTSIYLAVSPEVENVTGCYFADCQVKTLTETGRNTELARRLWEESEKIIAKK
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JSR3
A0A0N0PAY2
A0A2A4IXN8
A0A0L7KV18
I4DQA1
A0A0N0PB70
+ More
A0A2H1W6X7 A0A0N1ICN4 A0A1Q3G5H8 A0A0N0PAC1 A0A0N1IDL3 A0A212F5B7 A0A1Q3G5G7 A0A0L7KVJ8 A0A2A4ITG8 A0A2H1W7C7 A0A0N0PAN9 A0A3P8YKM0 A0A3P9A7S4 A0A1L8E4I5 A0A0N0PAY1 A0A3P8XDV8 A0A1L8E4K0 A0A0N1IF98 A0A2I4D8B2 W5NAH8 A0A3B3BDF8 A0A0L7L5N4 A0A3Q2UEG4 A0A2J7PGY1 A0A1Y1N8N2 A0A1Y1NCI4 K7FV47 H9GAK1 A0A3Q1J6H1 A0A3Q2DGZ5 A0A146XYN2 A0A147APK5 D6X0B0 H9JSL6 B3RKN4 A0A1A8VGE8 B5UAY9 W4ZIF1 H3AXM0 H2Z7K1 A0A3N0XRZ0 A0A0N0PAB9 B3RKN5 A0A1S3M3L0 A0A2D0PVD8 A0A060VRE0 A0A218UIJ9 A0A0N8E5H6 A0A1S3M424 A0A2J7PGT7 A0A1S3M363 A0A3P9JF09 A0A2J7PGT0 A0A0Q9WF80 A0A3P9I0S4 B3SEC9 B3SEC8 A0A0B7ANX9 B3RKK3 A0A1S3S0C8 A0A369RY25 A0A369RLX1 A0A1D2MFP1 B4LJG7 A0A0K8TLB0 H2N2X4 A0A3B3SMQ9 A0A3B3SLF6 A0A1B0GM47 A0A369RZQ8 A0A0M4EVS0 B4KPR0 A0A0Q9X8T9 A0A0P7XD55 A0A3B3SMR9 A0A1W5B461 A0A060XHT8 A0A3B3SMK7 A0A1S3QIK4 A0A0P5UI40 U3JRI9 A0A1S3JQV8 H2T513 A0A3B1JIS7 A0A0P8XPH1 A0A0Q3UTV3 A0A3Q3MS71 A0A2P6L863 A0A0P5YX32 A0A2I0U8W0 A0A0P6GLC1 A0A3Q3FY47 A0A3Q3MTU9 B0XLA9
A0A2H1W6X7 A0A0N1ICN4 A0A1Q3G5H8 A0A0N0PAC1 A0A0N1IDL3 A0A212F5B7 A0A1Q3G5G7 A0A0L7KVJ8 A0A2A4ITG8 A0A2H1W7C7 A0A0N0PAN9 A0A3P8YKM0 A0A3P9A7S4 A0A1L8E4I5 A0A0N0PAY1 A0A3P8XDV8 A0A1L8E4K0 A0A0N1IF98 A0A2I4D8B2 W5NAH8 A0A3B3BDF8 A0A0L7L5N4 A0A3Q2UEG4 A0A2J7PGY1 A0A1Y1N8N2 A0A1Y1NCI4 K7FV47 H9GAK1 A0A3Q1J6H1 A0A3Q2DGZ5 A0A146XYN2 A0A147APK5 D6X0B0 H9JSL6 B3RKN4 A0A1A8VGE8 B5UAY9 W4ZIF1 H3AXM0 H2Z7K1 A0A3N0XRZ0 A0A0N0PAB9 B3RKN5 A0A1S3M3L0 A0A2D0PVD8 A0A060VRE0 A0A218UIJ9 A0A0N8E5H6 A0A1S3M424 A0A2J7PGT7 A0A1S3M363 A0A3P9JF09 A0A2J7PGT0 A0A0Q9WF80 A0A3P9I0S4 B3SEC9 B3SEC8 A0A0B7ANX9 B3RKK3 A0A1S3S0C8 A0A369RY25 A0A369RLX1 A0A1D2MFP1 B4LJG7 A0A0K8TLB0 H2N2X4 A0A3B3SMQ9 A0A3B3SLF6 A0A1B0GM47 A0A369RZQ8 A0A0M4EVS0 B4KPR0 A0A0Q9X8T9 A0A0P7XD55 A0A3B3SMR9 A0A1W5B461 A0A060XHT8 A0A3B3SMK7 A0A1S3QIK4 A0A0P5UI40 U3JRI9 A0A1S3JQV8 H2T513 A0A3B1JIS7 A0A0P8XPH1 A0A0Q3UTV3 A0A3Q3MS71 A0A2P6L863 A0A0P5YX32 A0A2I0U8W0 A0A0P6GLC1 A0A3Q3FY47 A0A3Q3MTU9 B0XLA9
Pubmed
EMBL
BABH01032245
KQ461133
KPJ08901.1
NWSH01005060
PCG64471.1
JTDY01005329
+ More
KOB67107.1 AK404009 BAM20091.1 KPJ08899.1 ODYU01006737 SOQ48855.1 KQ458665 KPJ05597.1 GEYN01000183 JAV44806.1 KPJ05598.1 KPJ08900.1 AGBW02010210 OWR48927.1 GEYN01000196 JAV44793.1 KOB67106.1 NWSH01008708 PCG62440.1 SOQ48856.1 KPJ05596.1 GFDF01000434 JAV13650.1 KPJ08898.1 GFDF01000433 JAV13651.1 KPJ08897.1 AHAT01005059 JTDY01002853 KOB70599.1 NEVH01025161 PNF15592.1 GEZM01009786 JAV94304.1 GEZM01009785 JAV94305.1 AGCU01021282 AAWZ02022819 GCES01039838 JAR46485.1 GCES01005956 JAR80367.1 KQ971372 EFA10517.1 BABH01032240 DS985241 EDV29193.1 HAEJ01017786 SBS58243.1 AB439581 BAG72431.1 AAGJ04078790 AFYH01129353 RJVU01062584 ROJ29328.1 KPJ05595.1 EDV29194.1 FR904286 CDQ57371.1 MUZQ01000282 OWK53555.1 GDIQ01059860 JAN34877.1 PNF15542.1 PNF15543.1 CH940648 KRF80052.1 DS985430 EDV18917.1 EDV18916.1 HACG01034750 CEK81615.1 EDV29907.1 NOWV01000138 RDD39359.1 NOWV01000972 RDD35818.1 LJIJ01001415 ODM91806.1 EDW61535.1 GDAI01002635 JAI14968.1 AJVK01011173 NOWV01000110 RDD40168.1 CP012524 ALC42025.1 CH933808 EDW09170.2 KRG04501.1 JARO02002249 KPP73165.1 FR905416 CDQ79168.1 GDIP01112562 JAL91152.1 AGTO01010438 CH902619 KPU76518.1 LMAW01000825 KQK85306.1 MWRG01001082 PRD34763.1 GDIP01052070 JAM51645.1 KZ505992 PKU42472.1 GDIQ01042179 JAN52558.1 DS234386 EDS33797.1
KOB67107.1 AK404009 BAM20091.1 KPJ08899.1 ODYU01006737 SOQ48855.1 KQ458665 KPJ05597.1 GEYN01000183 JAV44806.1 KPJ05598.1 KPJ08900.1 AGBW02010210 OWR48927.1 GEYN01000196 JAV44793.1 KOB67106.1 NWSH01008708 PCG62440.1 SOQ48856.1 KPJ05596.1 GFDF01000434 JAV13650.1 KPJ08898.1 GFDF01000433 JAV13651.1 KPJ08897.1 AHAT01005059 JTDY01002853 KOB70599.1 NEVH01025161 PNF15592.1 GEZM01009786 JAV94304.1 GEZM01009785 JAV94305.1 AGCU01021282 AAWZ02022819 GCES01039838 JAR46485.1 GCES01005956 JAR80367.1 KQ971372 EFA10517.1 BABH01032240 DS985241 EDV29193.1 HAEJ01017786 SBS58243.1 AB439581 BAG72431.1 AAGJ04078790 AFYH01129353 RJVU01062584 ROJ29328.1 KPJ05595.1 EDV29194.1 FR904286 CDQ57371.1 MUZQ01000282 OWK53555.1 GDIQ01059860 JAN34877.1 PNF15542.1 PNF15543.1 CH940648 KRF80052.1 DS985430 EDV18917.1 EDV18916.1 HACG01034750 CEK81615.1 EDV29907.1 NOWV01000138 RDD39359.1 NOWV01000972 RDD35818.1 LJIJ01001415 ODM91806.1 EDW61535.1 GDAI01002635 JAI14968.1 AJVK01011173 NOWV01000110 RDD40168.1 CP012524 ALC42025.1 CH933808 EDW09170.2 KRG04501.1 JARO02002249 KPP73165.1 FR905416 CDQ79168.1 GDIP01112562 JAL91152.1 AGTO01010438 CH902619 KPU76518.1 LMAW01000825 KQK85306.1 MWRG01001082 PRD34763.1 GDIP01052070 JAM51645.1 KZ505992 PKU42472.1 GDIQ01042179 JAN52558.1 DS234386 EDS33797.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000037510
UP000053268
UP000007151
+ More
UP000265140 UP000192220 UP000018468 UP000261560 UP000265000 UP000235965 UP000007267 UP000001646 UP000265040 UP000265020 UP000007266 UP000009022 UP000007110 UP000008672 UP000007875 UP000087266 UP000221080 UP000193380 UP000197619 UP000265200 UP000008792 UP000253843 UP000094527 UP000001038 UP000261540 UP000092462 UP000092553 UP000009192 UP000034805 UP000192224 UP000016665 UP000085678 UP000005226 UP000018467 UP000007801 UP000051836 UP000261640 UP000261660 UP000002320
UP000265140 UP000192220 UP000018468 UP000261560 UP000265000 UP000235965 UP000007267 UP000001646 UP000265040 UP000265020 UP000007266 UP000009022 UP000007110 UP000008672 UP000007875 UP000087266 UP000221080 UP000193380 UP000197619 UP000265200 UP000008792 UP000253843 UP000094527 UP000001038 UP000261540 UP000092462 UP000092553 UP000009192 UP000034805 UP000192224 UP000016665 UP000085678 UP000005226 UP000018467 UP000007801 UP000051836 UP000261640 UP000261660 UP000002320
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JSR3
A0A0N0PAY2
A0A2A4IXN8
A0A0L7KV18
I4DQA1
A0A0N0PB70
+ More
A0A2H1W6X7 A0A0N1ICN4 A0A1Q3G5H8 A0A0N0PAC1 A0A0N1IDL3 A0A212F5B7 A0A1Q3G5G7 A0A0L7KVJ8 A0A2A4ITG8 A0A2H1W7C7 A0A0N0PAN9 A0A3P8YKM0 A0A3P9A7S4 A0A1L8E4I5 A0A0N0PAY1 A0A3P8XDV8 A0A1L8E4K0 A0A0N1IF98 A0A2I4D8B2 W5NAH8 A0A3B3BDF8 A0A0L7L5N4 A0A3Q2UEG4 A0A2J7PGY1 A0A1Y1N8N2 A0A1Y1NCI4 K7FV47 H9GAK1 A0A3Q1J6H1 A0A3Q2DGZ5 A0A146XYN2 A0A147APK5 D6X0B0 H9JSL6 B3RKN4 A0A1A8VGE8 B5UAY9 W4ZIF1 H3AXM0 H2Z7K1 A0A3N0XRZ0 A0A0N0PAB9 B3RKN5 A0A1S3M3L0 A0A2D0PVD8 A0A060VRE0 A0A218UIJ9 A0A0N8E5H6 A0A1S3M424 A0A2J7PGT7 A0A1S3M363 A0A3P9JF09 A0A2J7PGT0 A0A0Q9WF80 A0A3P9I0S4 B3SEC9 B3SEC8 A0A0B7ANX9 B3RKK3 A0A1S3S0C8 A0A369RY25 A0A369RLX1 A0A1D2MFP1 B4LJG7 A0A0K8TLB0 H2N2X4 A0A3B3SMQ9 A0A3B3SLF6 A0A1B0GM47 A0A369RZQ8 A0A0M4EVS0 B4KPR0 A0A0Q9X8T9 A0A0P7XD55 A0A3B3SMR9 A0A1W5B461 A0A060XHT8 A0A3B3SMK7 A0A1S3QIK4 A0A0P5UI40 U3JRI9 A0A1S3JQV8 H2T513 A0A3B1JIS7 A0A0P8XPH1 A0A0Q3UTV3 A0A3Q3MS71 A0A2P6L863 A0A0P5YX32 A0A2I0U8W0 A0A0P6GLC1 A0A3Q3FY47 A0A3Q3MTU9 B0XLA9
A0A2H1W6X7 A0A0N1ICN4 A0A1Q3G5H8 A0A0N0PAC1 A0A0N1IDL3 A0A212F5B7 A0A1Q3G5G7 A0A0L7KVJ8 A0A2A4ITG8 A0A2H1W7C7 A0A0N0PAN9 A0A3P8YKM0 A0A3P9A7S4 A0A1L8E4I5 A0A0N0PAY1 A0A3P8XDV8 A0A1L8E4K0 A0A0N1IF98 A0A2I4D8B2 W5NAH8 A0A3B3BDF8 A0A0L7L5N4 A0A3Q2UEG4 A0A2J7PGY1 A0A1Y1N8N2 A0A1Y1NCI4 K7FV47 H9GAK1 A0A3Q1J6H1 A0A3Q2DGZ5 A0A146XYN2 A0A147APK5 D6X0B0 H9JSL6 B3RKN4 A0A1A8VGE8 B5UAY9 W4ZIF1 H3AXM0 H2Z7K1 A0A3N0XRZ0 A0A0N0PAB9 B3RKN5 A0A1S3M3L0 A0A2D0PVD8 A0A060VRE0 A0A218UIJ9 A0A0N8E5H6 A0A1S3M424 A0A2J7PGT7 A0A1S3M363 A0A3P9JF09 A0A2J7PGT0 A0A0Q9WF80 A0A3P9I0S4 B3SEC9 B3SEC8 A0A0B7ANX9 B3RKK3 A0A1S3S0C8 A0A369RY25 A0A369RLX1 A0A1D2MFP1 B4LJG7 A0A0K8TLB0 H2N2X4 A0A3B3SMQ9 A0A3B3SLF6 A0A1B0GM47 A0A369RZQ8 A0A0M4EVS0 B4KPR0 A0A0Q9X8T9 A0A0P7XD55 A0A3B3SMR9 A0A1W5B461 A0A060XHT8 A0A3B3SMK7 A0A1S3QIK4 A0A0P5UI40 U3JRI9 A0A1S3JQV8 H2T513 A0A3B1JIS7 A0A0P8XPH1 A0A0Q3UTV3 A0A3Q3MS71 A0A2P6L863 A0A0P5YX32 A0A2I0U8W0 A0A0P6GLC1 A0A3Q3FY47 A0A3Q3MTU9 B0XLA9
PDB
3RD5
E-value=1.14682e-26,
Score=296
Ontologies
KEGG
PATHWAY
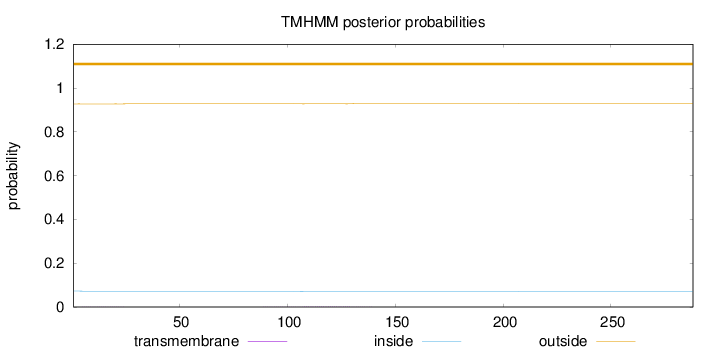
Topology
Length:
288
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05278
Exp number, first 60 AAs:
0.01043
Total prob of N-in:
0.07254
outside
1 - 288
Population Genetic Test Statistics
Pi
230.878971
Theta
164.613685
Tajima's D
1.266022
CLR
0
CSRT
0.729713514324284
Interpretation
Uncertain
