Gene
KWMTBOMO05019
Pre Gene Modal
BGIBMGA012526
Annotation
PREDICTED:_alanine--glyoxylate_aminotransferase_2-like_[Papilio_machaon]
Full name
Alanine--glyoxylate aminotransferase 2-like
Location in the cell
Cytoplasmic Reliability : 3.183
Sequence
CDS
ATGGCTTATTTACAATCAATGCCCAAATCTGAAACCATTCAACTTCGCGAGAAACATGTTGGAGCGGCGTGCCAACTGTTTTTCCGTTCTTCCCCCTTGAAAATAGTGCGAGGAATAGCACAGTTCATGTATGACGAAACCGGCGAAAGGTATCTCGACTGCATCAACAACGTTGCCCATGTCGGCCATTGTCATCCACATGTTGTTGAGGCTGGTAGAAATCAAATGTCTCTAATATCGACAAACAACCGCTATCTTCACGACGAGCTAGTTATTCTAGCACAAAGGCTCGTGAATACATTACCAGAATCATTGTCCGTTTGCTTCTTCGTAAACTCTGGGTCGGAGGCTAATGATCTCGCTTTGAGAATGGCAAGAATCCATACTAAGAAGAAGGATGTAATTACCTTGGACCACGCGTATCATGGCCATCTAACGACTATGATCGATATTTCACCATACAAATTTAACCTACCCGGAGGTCCCGAGAAACCAGACTGGGTTCATGTGGCACCAGTACCGGACGTGTACAGGGGGAAATATACTCATCCGGCGGACTCAGAATCGGTAGAGGAAGTAGGTCGGATGTACGCCGACGAAGTAGGCAATATCTGCAATGAGATTAAGAATAAGAATGGAGGCGTTTGTGCTTTCATAGCGGAGAGTCTACAGAGCTGTGGGGGACAGATTATACCTCCGGACGGATATTTCAAACGTGTATACGAATATGTGCATGAAGCCGGTGGTGTGTGTATCGCCGATGAGGTTCAAGTAGGGTTCGGTCGTGTCGGCACACACATGTGGGCCTTTGAAACTCAAGACGTTGTTCCCGACATAGTCACTATGGGCAAACCTATGGGTAATGGACATCCTGTGGCCGCTGTAATTACCACCCCCGAGATCGCCAAGAGCTTCTCTGATACAGGCGTTGAATATTTTAACACGTACGGCGGCAATCCTGTGTCTTGTGCCATCGCCAATGCAGTACTTGATGTCATCGAAGAAGAAAACCTTTTGGAGAGAGCGTCACGTGTCGGTAACCATTTACTCAGTCGGTGTGAAGATCTGAAGCATAAACATAGACTCGTCGGCGATGTCCGCGGTCGGGGTCTGTTTGTAGGCGTCGAACTTGTCACCGACCGGGAAACCAGAACTCCCGCTACAGCTGAAGCAAAACATGTTGTCAATAGAATGAGAGAAGAAAATATATTGATTAGTCGTGACGGGCCCGACAGTAACGTATTAAAGTTTAAGCCCCCCATGGTGTTTACAACACAAGACGCCGACAGACTAGTGGAAACTCTAGACAGGGTTTTAGGGGAGTTGGATGACACAAGAATATCGAATATTAAATTGGAAGTACTAGTCACACCGATCAACACTGAAGTTCCGAAAAAGGAGAACGTCATTAAAAGTTCTGTACAAAAGGCAATCAAGACTTGA
Protein
MAYLQSMPKSETIQLREKHVGAACQLFFRSSPLKIVRGIAQFMYDETGERYLDCINNVAHVGHCHPHVVEAGRNQMSLISTNNRYLHDELVILAQRLVNTLPESLSVCFFVNSGSEANDLALRMARIHTKKKDVITLDHAYHGHLTTMIDISPYKFNLPGGPEKPDWVHVAPVPDVYRGKYTHPADSESVEEVGRMYADEVGNICNEIKNKNGGVCAFIAESLQSCGGQIIPPDGYFKRVYEYVHEAGGVCIADEVQVGFGRVGTHMWAFETQDVVPDIVTMGKPMGNGHPVAAVITTPEIAKSFSDTGVEYFNTYGGNPVSCAIANAVLDVIEEENLLERASRVGNHLLSRCEDLKHKHRLVGDVRGRGLFVGVELVTDRETRTPATAEAKHVVNRMREENILISRDGPDSNVLKFKPPMVFTTQDADRLVETLDRVLGELDDTRISNIKLEVLVTPINTEVPKKENVIKSSVQKAIKT
Summary
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the class-III pyridoxal-phosphate-dependent aminotransferase family.
Keywords
Complete proteome
Lyase
Pyridoxal phosphate
Reference proteome
Feature
chain Alanine--glyoxylate aminotransferase 2-like
Uniprot
H9JSL5
A0A194QR94
A0A2A4IUP8
A0A0N1PHP5
A0A2H1W7N4
A0A212EVU3
+ More
S4PE92 A0A3S2N7Q6 A0A026WAC1 A0A3L8DY65 A0A151I060 A0A195DE54 E2ANT3 A0A2A3EMQ9 V9IJY5 A0A088ADL7 A0A154NYL6 A0A158NYL7 E9JDG7 A0A195FV90 F4WXZ5 D6X4E0 A0A1W4WGP5 A0A151IEU4 A0A151WUJ8 A0A2J7PYD2 A0A2J7PYC9 A0A1Y1KIK8 K7J0R3 A0A1B6E588 A0A1S3H3Y7 U4UQ15 A0A1D2MQK8 A0A0K8TUA3 A0A182QMT5 A0A0N7ZCN1 B4JWU9 A0A182NC35 A0A2J7PYB8 A0A034VHP8 A0A0K8TYF0 A0A0K8USD2 A0A182XBU8 A0A182IAN7 A0A023EVX0 A0A0N8C8M0 Q7QF58 B0WHU8 A0A182USM3 A0A0A1WVH8 A0A0N8BX71 A0A182TRB9 A0A182GH95 A0A0P6E6F6 A0A0P6BDG1 A0A0N8EHH5 A0A2M4AAD8 B4M307 Q16M11 T1DJ34 A0A1B6FGY7 A0A1A9V726 E9HCX2 W8B8Q8 A0A2M4AAQ4 A0A0P5J5B2 A0A182FJ56 A0A1Q3FQD5 W5JHY9 B4L5J3 A0A182MDZ1 A0A1I8QEQ9 A0A2M3ZGU4 A0A2M4BMA2 A0A1B6KV85 A0A2M3Z365 A0A2M3Z3E5 Q0QHL6 B3MZU8 A0A0P5RAP5 A0A182RM56 A0A0M4FAZ4 A0A1L8E276 A0A3B0KEN6 B4N0Z8 B5DN06 A0A1J1HHA1 B4QJH1 A0A1I8N1F2 B3NHH6 A0A1A9W8J3 Q9VU95 A0A2S2NDT5 A0A0P5IQW3 B4PHW5 R7VFZ7 A0A1W4UII8 A0A0N8C3X5 J9K0G9 T1JKM4
S4PE92 A0A3S2N7Q6 A0A026WAC1 A0A3L8DY65 A0A151I060 A0A195DE54 E2ANT3 A0A2A3EMQ9 V9IJY5 A0A088ADL7 A0A154NYL6 A0A158NYL7 E9JDG7 A0A195FV90 F4WXZ5 D6X4E0 A0A1W4WGP5 A0A151IEU4 A0A151WUJ8 A0A2J7PYD2 A0A2J7PYC9 A0A1Y1KIK8 K7J0R3 A0A1B6E588 A0A1S3H3Y7 U4UQ15 A0A1D2MQK8 A0A0K8TUA3 A0A182QMT5 A0A0N7ZCN1 B4JWU9 A0A182NC35 A0A2J7PYB8 A0A034VHP8 A0A0K8TYF0 A0A0K8USD2 A0A182XBU8 A0A182IAN7 A0A023EVX0 A0A0N8C8M0 Q7QF58 B0WHU8 A0A182USM3 A0A0A1WVH8 A0A0N8BX71 A0A182TRB9 A0A182GH95 A0A0P6E6F6 A0A0P6BDG1 A0A0N8EHH5 A0A2M4AAD8 B4M307 Q16M11 T1DJ34 A0A1B6FGY7 A0A1A9V726 E9HCX2 W8B8Q8 A0A2M4AAQ4 A0A0P5J5B2 A0A182FJ56 A0A1Q3FQD5 W5JHY9 B4L5J3 A0A182MDZ1 A0A1I8QEQ9 A0A2M3ZGU4 A0A2M4BMA2 A0A1B6KV85 A0A2M3Z365 A0A2M3Z3E5 Q0QHL6 B3MZU8 A0A0P5RAP5 A0A182RM56 A0A0M4FAZ4 A0A1L8E276 A0A3B0KEN6 B4N0Z8 B5DN06 A0A1J1HHA1 B4QJH1 A0A1I8N1F2 B3NHH6 A0A1A9W8J3 Q9VU95 A0A2S2NDT5 A0A0P5IQW3 B4PHW5 R7VFZ7 A0A1W4UII8 A0A0N8C3X5 J9K0G9 T1JKM4
EC Number
4.2.3.-
Pubmed
19121390
26354079
22118469
23622113
24508170
30249741
+ More
20798317 21347285 21282665 21719571 18362917 19820115 28004739 20075255 23537049 27289101 26369729 17994087 25348373 24945155 12364791 14747013 17210077 25830018 26483478 17510324 21292972 24495485 20920257 23761445 20353571 15632085 22936249 25315136 10731132 12537572 12537569 17550304 23254933
20798317 21347285 21282665 21719571 18362917 19820115 28004739 20075255 23537049 27289101 26369729 17994087 25348373 24945155 12364791 14747013 17210077 25830018 26483478 17510324 21292972 24495485 20920257 23761445 20353571 15632085 22936249 25315136 10731132 12537572 12537569 17550304 23254933
EMBL
BABH01032250
KQ461198
KPJ06056.1
NWSH01006063
PCG63697.1
KQ458665
+ More
KPJ05600.1 ODYU01006737 SOQ48852.1 AGBW02012137 OWR45577.1 GAIX01004627 JAA87933.1 RSAL01000238 RVE43702.1 KK107295 EZA53000.1 QOIP01000003 RLU25232.1 KQ976631 KYM78899.1 KQ980953 KYN11166.1 GL441336 EFN64903.1 KZ288206 PBC33093.1 JR051218 AEY61453.1 KQ434773 KZC03970.1 ADTU01004045 GL771866 EFZ09126.1 KQ981276 KYN43784.1 GL888434 EGI60931.1 KQ971410 EEZ97540.1 KQ977854 KYM99342.1 KQ982748 KYQ51315.1 NEVH01020850 PNF21322.1 PNF21324.1 GEZM01087540 JAV58627.1 GEDC01004231 JAS33067.1 KB632410 ERL95192.1 LJIJ01000681 ODM95367.1 GDAI01000078 JAI17525.1 AXCN02001428 GDRN01064003 GDRN01064001 JAI64912.1 CH916376 EDV95225.1 PNF21328.1 GAKP01016919 GAKP01016918 JAC42034.1 GDHF01033028 JAI19286.1 GDHF01022823 JAI29491.1 APCN01001282 GAPW01001139 JAC12459.1 GDIQ01169678 GDIQ01120397 GDIQ01104099 GDIQ01080457 JAL47627.1 AAAB01008846 EAA06360.5 DS231940 EDS28007.1 GBXI01011757 JAD02535.1 GDIQ01136091 JAL15635.1 JXUM01001293 KQ560119 KXJ84391.1 GDIQ01067224 JAN27513.1 GDIP01018665 JAM85050.1 GDIQ01026268 JAN68469.1 GGFK01004404 MBW37725.1 CH940651 EDW65182.1 CH477884 EAT35372.1 GAMD01001651 JAA99939.1 GECZ01020375 JAS49394.1 GL732621 EFX70448.1 GAMC01008900 JAB97655.1 GGFK01004397 MBW37718.1 GDIQ01205061 JAK46664.1 GFDL01005240 JAV29805.1 ADMH02001354 ETN62898.1 CH933811 EDW06452.1 AXCM01005244 GGFM01006907 MBW27658.1 GGFJ01004757 MBW53898.1 GEBQ01024634 JAT15343.1 GGFM01002211 MBW22962.1 GGFM01002274 MBW23025.1 DQ374003 EZ423984 ABG77981.1 ADD20260.1 CH902635 EDV33899.1 GDIQ01122997 JAL28729.1 CP012528 ALC49682.1 GFDF01001244 JAV12840.1 OUUW01000011 SPP86790.1 CH963920 EDW78160.1 CH379064 EDY72813.1 CVRI01000004 CRK87381.1 CM000363 CM002912 EDX10380.1 KMY99467.1 CH954178 EDV51702.1 AE014296 AY061111 GGMR01002676 MBY15295.1 GDIQ01211616 JAK40109.1 CM000159 EDW94440.1 AMQN01016708 KB292329 ELU17763.1 GDIQ01117259 JAL34467.1 ABLF02036955 ABLF02036967 JH431720
KPJ05600.1 ODYU01006737 SOQ48852.1 AGBW02012137 OWR45577.1 GAIX01004627 JAA87933.1 RSAL01000238 RVE43702.1 KK107295 EZA53000.1 QOIP01000003 RLU25232.1 KQ976631 KYM78899.1 KQ980953 KYN11166.1 GL441336 EFN64903.1 KZ288206 PBC33093.1 JR051218 AEY61453.1 KQ434773 KZC03970.1 ADTU01004045 GL771866 EFZ09126.1 KQ981276 KYN43784.1 GL888434 EGI60931.1 KQ971410 EEZ97540.1 KQ977854 KYM99342.1 KQ982748 KYQ51315.1 NEVH01020850 PNF21322.1 PNF21324.1 GEZM01087540 JAV58627.1 GEDC01004231 JAS33067.1 KB632410 ERL95192.1 LJIJ01000681 ODM95367.1 GDAI01000078 JAI17525.1 AXCN02001428 GDRN01064003 GDRN01064001 JAI64912.1 CH916376 EDV95225.1 PNF21328.1 GAKP01016919 GAKP01016918 JAC42034.1 GDHF01033028 JAI19286.1 GDHF01022823 JAI29491.1 APCN01001282 GAPW01001139 JAC12459.1 GDIQ01169678 GDIQ01120397 GDIQ01104099 GDIQ01080457 JAL47627.1 AAAB01008846 EAA06360.5 DS231940 EDS28007.1 GBXI01011757 JAD02535.1 GDIQ01136091 JAL15635.1 JXUM01001293 KQ560119 KXJ84391.1 GDIQ01067224 JAN27513.1 GDIP01018665 JAM85050.1 GDIQ01026268 JAN68469.1 GGFK01004404 MBW37725.1 CH940651 EDW65182.1 CH477884 EAT35372.1 GAMD01001651 JAA99939.1 GECZ01020375 JAS49394.1 GL732621 EFX70448.1 GAMC01008900 JAB97655.1 GGFK01004397 MBW37718.1 GDIQ01205061 JAK46664.1 GFDL01005240 JAV29805.1 ADMH02001354 ETN62898.1 CH933811 EDW06452.1 AXCM01005244 GGFM01006907 MBW27658.1 GGFJ01004757 MBW53898.1 GEBQ01024634 JAT15343.1 GGFM01002211 MBW22962.1 GGFM01002274 MBW23025.1 DQ374003 EZ423984 ABG77981.1 ADD20260.1 CH902635 EDV33899.1 GDIQ01122997 JAL28729.1 CP012528 ALC49682.1 GFDF01001244 JAV12840.1 OUUW01000011 SPP86790.1 CH963920 EDW78160.1 CH379064 EDY72813.1 CVRI01000004 CRK87381.1 CM000363 CM002912 EDX10380.1 KMY99467.1 CH954178 EDV51702.1 AE014296 AY061111 GGMR01002676 MBY15295.1 GDIQ01211616 JAK40109.1 CM000159 EDW94440.1 AMQN01016708 KB292329 ELU17763.1 GDIQ01117259 JAL34467.1 ABLF02036955 ABLF02036967 JH431720
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000283053
+ More
UP000053097 UP000279307 UP000078540 UP000078492 UP000000311 UP000242457 UP000005203 UP000076502 UP000005205 UP000078541 UP000007755 UP000007266 UP000192223 UP000078542 UP000075809 UP000235965 UP000002358 UP000085678 UP000030742 UP000094527 UP000075886 UP000001070 UP000075884 UP000076407 UP000075840 UP000007062 UP000002320 UP000075903 UP000075902 UP000069940 UP000249989 UP000008792 UP000008820 UP000078200 UP000000305 UP000069272 UP000000673 UP000009192 UP000075883 UP000095300 UP000007801 UP000075900 UP000092553 UP000268350 UP000007798 UP000001819 UP000183832 UP000000304 UP000095301 UP000008711 UP000091820 UP000000803 UP000002282 UP000014760 UP000192221 UP000007819
UP000053097 UP000279307 UP000078540 UP000078492 UP000000311 UP000242457 UP000005203 UP000076502 UP000005205 UP000078541 UP000007755 UP000007266 UP000192223 UP000078542 UP000075809 UP000235965 UP000002358 UP000085678 UP000030742 UP000094527 UP000075886 UP000001070 UP000075884 UP000076407 UP000075840 UP000007062 UP000002320 UP000075903 UP000075902 UP000069940 UP000249989 UP000008792 UP000008820 UP000078200 UP000000305 UP000069272 UP000000673 UP000009192 UP000075883 UP000095300 UP000007801 UP000075900 UP000092553 UP000268350 UP000007798 UP000001819 UP000183832 UP000000304 UP000095301 UP000008711 UP000091820 UP000000803 UP000002282 UP000014760 UP000192221 UP000007819
Interpro
IPR015421
PyrdxlP-dep_Trfase_major
+ More
IPR015424 PyrdxlP-dep_Trfase
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR005814 Aminotrans_3
IPR011030 Lipovitellin_superhlx_dom
IPR001846 VWF_type-D
IPR001747 Lipid_transpt_N
IPR015819 Lipid_transp_b-sht_shell
IPR015816 Vitellinogen_b-sht_N
IPR015255 Vitellinogen_open_b-sht
IPR015424 PyrdxlP-dep_Trfase
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR005814 Aminotrans_3
IPR011030 Lipovitellin_superhlx_dom
IPR001846 VWF_type-D
IPR001747 Lipid_transpt_N
IPR015819 Lipid_transp_b-sht_shell
IPR015816 Vitellinogen_b-sht_N
IPR015255 Vitellinogen_open_b-sht
Gene 3D
CDD
ProteinModelPortal
H9JSL5
A0A194QR94
A0A2A4IUP8
A0A0N1PHP5
A0A2H1W7N4
A0A212EVU3
+ More
S4PE92 A0A3S2N7Q6 A0A026WAC1 A0A3L8DY65 A0A151I060 A0A195DE54 E2ANT3 A0A2A3EMQ9 V9IJY5 A0A088ADL7 A0A154NYL6 A0A158NYL7 E9JDG7 A0A195FV90 F4WXZ5 D6X4E0 A0A1W4WGP5 A0A151IEU4 A0A151WUJ8 A0A2J7PYD2 A0A2J7PYC9 A0A1Y1KIK8 K7J0R3 A0A1B6E588 A0A1S3H3Y7 U4UQ15 A0A1D2MQK8 A0A0K8TUA3 A0A182QMT5 A0A0N7ZCN1 B4JWU9 A0A182NC35 A0A2J7PYB8 A0A034VHP8 A0A0K8TYF0 A0A0K8USD2 A0A182XBU8 A0A182IAN7 A0A023EVX0 A0A0N8C8M0 Q7QF58 B0WHU8 A0A182USM3 A0A0A1WVH8 A0A0N8BX71 A0A182TRB9 A0A182GH95 A0A0P6E6F6 A0A0P6BDG1 A0A0N8EHH5 A0A2M4AAD8 B4M307 Q16M11 T1DJ34 A0A1B6FGY7 A0A1A9V726 E9HCX2 W8B8Q8 A0A2M4AAQ4 A0A0P5J5B2 A0A182FJ56 A0A1Q3FQD5 W5JHY9 B4L5J3 A0A182MDZ1 A0A1I8QEQ9 A0A2M3ZGU4 A0A2M4BMA2 A0A1B6KV85 A0A2M3Z365 A0A2M3Z3E5 Q0QHL6 B3MZU8 A0A0P5RAP5 A0A182RM56 A0A0M4FAZ4 A0A1L8E276 A0A3B0KEN6 B4N0Z8 B5DN06 A0A1J1HHA1 B4QJH1 A0A1I8N1F2 B3NHH6 A0A1A9W8J3 Q9VU95 A0A2S2NDT5 A0A0P5IQW3 B4PHW5 R7VFZ7 A0A1W4UII8 A0A0N8C3X5 J9K0G9 T1JKM4
S4PE92 A0A3S2N7Q6 A0A026WAC1 A0A3L8DY65 A0A151I060 A0A195DE54 E2ANT3 A0A2A3EMQ9 V9IJY5 A0A088ADL7 A0A154NYL6 A0A158NYL7 E9JDG7 A0A195FV90 F4WXZ5 D6X4E0 A0A1W4WGP5 A0A151IEU4 A0A151WUJ8 A0A2J7PYD2 A0A2J7PYC9 A0A1Y1KIK8 K7J0R3 A0A1B6E588 A0A1S3H3Y7 U4UQ15 A0A1D2MQK8 A0A0K8TUA3 A0A182QMT5 A0A0N7ZCN1 B4JWU9 A0A182NC35 A0A2J7PYB8 A0A034VHP8 A0A0K8TYF0 A0A0K8USD2 A0A182XBU8 A0A182IAN7 A0A023EVX0 A0A0N8C8M0 Q7QF58 B0WHU8 A0A182USM3 A0A0A1WVH8 A0A0N8BX71 A0A182TRB9 A0A182GH95 A0A0P6E6F6 A0A0P6BDG1 A0A0N8EHH5 A0A2M4AAD8 B4M307 Q16M11 T1DJ34 A0A1B6FGY7 A0A1A9V726 E9HCX2 W8B8Q8 A0A2M4AAQ4 A0A0P5J5B2 A0A182FJ56 A0A1Q3FQD5 W5JHY9 B4L5J3 A0A182MDZ1 A0A1I8QEQ9 A0A2M3ZGU4 A0A2M4BMA2 A0A1B6KV85 A0A2M3Z365 A0A2M3Z3E5 Q0QHL6 B3MZU8 A0A0P5RAP5 A0A182RM56 A0A0M4FAZ4 A0A1L8E276 A0A3B0KEN6 B4N0Z8 B5DN06 A0A1J1HHA1 B4QJH1 A0A1I8N1F2 B3NHH6 A0A1A9W8J3 Q9VU95 A0A2S2NDT5 A0A0P5IQW3 B4PHW5 R7VFZ7 A0A1W4UII8 A0A0N8C3X5 J9K0G9 T1JKM4
PDB
5G4J
E-value=1.37799e-78,
Score=747
Ontologies
PATHWAY
GO
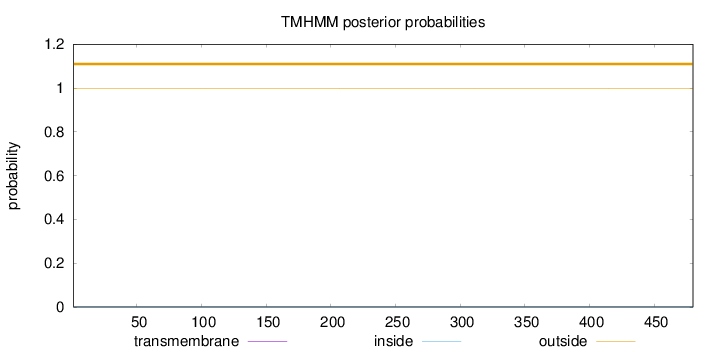
Topology
Length:
480
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00305
Exp number, first 60 AAs:
0.00145
Total prob of N-in:
0.00324
outside
1 - 480
Population Genetic Test Statistics
Pi
198.80119
Theta
166.159446
Tajima's D
0.065975
CLR
1.019841
CSRT
0.393130343482826
Interpretation
Uncertain
