Pre Gene Modal
BGIBMGA012577
Annotation
PREDICTED:_probable_E3_ubiquitin-protein_ligase_HERC4_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.325
Sequence
CDS
ATGTTTTGTTGGGGCAATGCAACTCATCATGAACTGTGCATCGACGGTTCACAAATAACTGATTTGGTTATCAAGCCCACATTATCAAAATGGAAAGAAAGTGATCACATACAAGCAGTAGCAGCTGGCGAATTCCATAGCTTATACCTCTCAAACAGTGGTCATTTATATTCATGTGGAAATAATGATGTGGGTCAGTTAGGACGTCAAACTGAATCTGACAATGGAAAGAATCCAGCTTTAGTGGAAACATTTAAAGACTGCACAATATCGACAATAGCATGTGGCCTTCAACATTCTTTAGCTATAGATGAATGGGGGCAGCCTTTCAGTTGGGGCTCTGACAGTATGGGGCAACTTGGCAATAACCTTGGCGCTCATGCTCAGGACAAACCTAAATTTGTCAAAGGGTTAGCAACCAAAAATGTAATACAAGTAGCTTGTGGAGCCTATCATTCTGTGGCATTAACAAACAGTGGTGATCTCTACTCATGGGGTGCAAATAGTTATGGACAATGTGGTTTAGGAACTATATCAAACAAGGAAATGTCACCTCAACACATTTCATCATTAATTGGTGTACCTATTGCCTTAATAGCATGTGGCAATAACCATACATTTGTGCTATCAAAATCAGGTGCGGTTTTTGGATGGGGTAAAAACACTCATGGACAACTTGGTCTTCAAGACGTTGAAAACAAATGCTATCCCACTCACCTAAAAACATTGAGAAATGTGAAGGTATGTCACATAGCGTGTGGTGAAGATTTCACTGTTTTCTTGACATTGGACGGTGGTGTGTTCACATGTGGAACTGGTGAATACGGGCAGACTGGACATGGTACGACTAGAGATGAGCTTGTTCCAAGAAAGATTGTCGAGCTGATGGGCAGTACAGTGACGCAGATAGCGTGCGGACGACGGCACTTGCTGTGTCGCGTCGGCGAGCGAATCCTGGCTTGCGGTTACGGAGCGCGAGGTCAACTTGGCTGTCCGCACATGGCGTTCGCGCTGGTCCCGACACCTGTACCGTTCACACCCAACGACGAATCTCCGTTTTCACCTGAAATCTTCAAGGGTCCGATTAAAGTGTTCGCGGGAGGTGACCACAGCTTCGTGGTATTGAAAGGCGACAAGTCCGGGCCGGTCGACTACCGCACTCCGGACCACAGGAAGCAAATACTCACGCTCACTCTACCGAAGCTAGCCGCGTGTCAAGTCTTCAAAGATAAAGATTTCGTTAATCAGGATTTAATGGCGTACTTAGAGACGGTGTTCGGTTCGCTGGCGTGTGTGAACGCGTCCTTCCTGCTGCCGAGCAACGGACACTACTGTTGCAACACCAAAATGCCGGGCGTCGACCTTAGCAAAGCCGAGGAGGCGTTCTCGTTGATCAGCCGATTCGAAAACACGTCCATCAAGGAACTAATATTCAGTCACTTAACGGATAATATAATTAAGAAAATGAAGCCCTCTCCACCGGATGCTGAAGCTCTAAGAATGTTTCTAATATTACCGCTATATCACGAGATGAGAAATCCCAGACGCCATACCCAGCTACAGGGTCCCTTTGCTGAAGCCTTCAACAACTTGTCTACTCATCCTCAAAAAATAGTCAACTTATGGTGGGAGGCGCAGACGACGGAGTACTTTGAGATGCTCGTAGATATATTCAAAAGCGTCGTTGTGTACGAACTTATGCAGCCTGTAGTTAGATCGACTAAGAAAGTGCAATTCAAACACAGCACCGTACAGATATTGAACACGTTGTCGTCGCTGAACAAGATCAACTTCACGAACCCGAAGCTACCTAAAATACCGGCTGAATGTTTCTACATAGAGGACTTGTGTGATCACGTTGACATCGCTTCTGATTATATTAACTGGTTGTCGGACCAAACTTCTACCCAGACGCACATGTGCAATTACGCTTTCCTGTTCGACGTCCAATGCAAGTCGTTGCTGCTGAAGATCGATCAGCAGCTACAAATGCAGATCGCCATAAGCAGGGCCACCACGCAAATCTTCACGAGGCTCTTCATGGATCCGACCTACGAGTATCAGAGGGATCAGTTCCTCAACCTGACCGTCTCGAGGAACCATATCGTCAGGGACACCATGCTGCAGATAAGCAACCATGACACTTCACAACTGAAGAAACCGCTCAGAGTCGAGTTTATTGGCGAAGAAGCGGAAGATGCCGGTGGCGTTAAAAAAGAATTCTTCATGCTACTGCTCAAGGAAATATTTGATCCCGTATACGGTATGTTCAAGCAATCCGAGGAGACGAACATGATTTGGTTTTCAAATAATCCATTCGAAGATGACGTCATGTATTATGTGATTGGGGCCATTTACGGACTTGCCATATACAATTCGATTATTATATACGTTCCGTTCCCGTTAGTTTTGTATAAGAAAATTCTCGGCGAATCTGTTATGATGGACGATTTGTCAGATTTGTATCCAACGTTAGCCAATAGTTTGAAAAGTTTATTGGAATATACAGACGATGACGTAGAGGAGGTGTTCGGACTGTGTTTCGCCGTGAACACTGAAGTGTTTGATCAAATACAAGTGAATCCACTGAAAGAGGACGGAGAGAATATACCTGTGACGCGCGAAAATAAAGCCGAATACGTTGAGTTGTACGTGGACTTTTTGCTCAATAAATCTGTGGAGAATCAATTTAAAGCTTTCAATCAAGGTTTCCAGAAGGTGTGCGGGGGTAGAATAATAAAGCTGTTTAGATCTCACGAATTGATGTCCGTTGTCATCGGAAACGAGGAGTACGACTGGGAAATGTTTCAGAACAATTGTGAATACAAGAATGGATACTCTGAAACTGATCAACAAATCAGATGGTTCTGGGAAACGTTCCACGAGCTGCCGCTTGAAGATAAGAAGAAATTTTTGCTTTTCTTAACCGGCAGCGATCGTGTTCCGATACAAGGAATGAGGGACATTAAAATAAGAATTCAACCAGTGGCAGACGACAGGTTCTTCCCGGTGGCCCACACTTGCTTCAATCTTCTAGATCTCCCGCGTTACAAGACTAAAGAGAGGCTCAAGTATTATCTTCTGCAGGCCATTCAGCAGACGCAGGGCTTCTCGCTGGTCTGA
Protein
MFCWGNATHHELCIDGSQITDLVIKPTLSKWKESDHIQAVAAGEFHSLYLSNSGHLYSCGNNDVGQLGRQTESDNGKNPALVETFKDCTISTIACGLQHSLAIDEWGQPFSWGSDSMGQLGNNLGAHAQDKPKFVKGLATKNVIQVACGAYHSVALTNSGDLYSWGANSYGQCGLGTISNKEMSPQHISSLIGVPIALIACGNNHTFVLSKSGAVFGWGKNTHGQLGLQDVENKCYPTHLKTLRNVKVCHIACGEDFTVFLTLDGGVFTCGTGEYGQTGHGTTRDELVPRKIVELMGSTVTQIACGRRHLLCRVGERILACGYGARGQLGCPHMAFALVPTPVPFTPNDESPFSPEIFKGPIKVFAGGDHSFVVLKGDKSGPVDYRTPDHRKQILTLTLPKLAACQVFKDKDFVNQDLMAYLETVFGSLACVNASFLLPSNGHYCCNTKMPGVDLSKAEEAFSLISRFENTSIKELIFSHLTDNIIKKMKPSPPDAEALRMFLILPLYHEMRNPRRHTQLQGPFAEAFNNLSTHPQKIVNLWWEAQTTEYFEMLVDIFKSVVVYELMQPVVRSTKKVQFKHSTVQILNTLSSLNKINFTNPKLPKIPAECFYIEDLCDHVDIASDYINWLSDQTSTQTHMCNYAFLFDVQCKSLLLKIDQQLQMQIAISRATTQIFTRLFMDPTYEYQRDQFLNLTVSRNHIVRDTMLQISNHDTSQLKKPLRVEFIGEEAEDAGGVKKEFFMLLLKEIFDPVYGMFKQSEETNMIWFSNNPFEDDVMYYVIGAIYGLAIYNSIIIYVPFPLVLYKKILGESVMMDDLSDLYPTLANSLKSLLEYTDDDVEEVFGLCFAVNTEVFDQIQVNPLKEDGENIPVTRENKAEYVELYVDFLLNKSVENQFKAFNQGFQKVCGGRIIKLFRSHELMSVVIGNEEYDWEMFQNNCEYKNGYSETDQQIRWFWETFHELPLEDKKKFLLFLTGSDRVPIQGMRDIKIRIQPVADDRFFPVAHTCFNLLDLPRYKTKERLKYYLLQAIQQTQGFSLV
Summary
Uniprot
H9JSR6
A0A2A4J1M4
A0A212EVR6
A0A0N1PGC6
A0A3S2P7H9
A0A194QKY1
+ More
A0A1W4X5Q9 D6W9I3 A0A2J7Q0N6 A0A1Y1NCD0 A0A2J7Q0N2 A0A1Y1NEM8 A0A067QLX8 E0VG83 A0A182V393 A0A182R9U6 A0A182P132 A0A2C9GS90 Q7PT00 A7URT7 A0A1B6FXJ2 A0A1B6ECJ7 A0A0K8SYD2 A0A0K8SYP3 A0A182Y317 A0A0K8SYD6 A0A1B6ME28 A0A1B6L8H2 A0A1Q3F9M7 A0A1B6G8R1 A0A1Q3F9S7 E2C7G3 A0A224XIS4 A0A182M4V1 A0A023F3C0 A0A182JTG2 A0A1B6KL25 B0X7X1 A0A1B6KIM6 A0A151JBP9 A0A0P4VZS4 A0A182ILN7 A0A084WRP8 A0A2M4AHB6 A0A182WWM0 A0A0V0G4T3 A0A182FGC0 E2AGG4 A0A2M4AHL9 A0A158NRC6 T1IG46 A0A182WHX5 F4WB37 A0A2M4CYM4 A0A1A9V478 A0A195FJJ8 A0A1I8PEI6 W5JA25 A0A088A9B2 A0A2A3EG75 A0A0J7KIS1 A0A151XF13 Q178R9 A0A151IFJ4 A0A1I8MYV6 A0A1I8PEQ1 A0A1I8MYV4 A0A182N9R9 A0A1Y1NBS2 A0A0L7RJK0 A0A336LTB2 A0A0K8V8N7 A0A1A9WLI1 K7JBL0 A0A1B6DL92 A0A0L0BV94 W8B7F6 A0A0C9QA00 A0A0C9R532 A0A232EEZ9 A0A034VTP6 A0A026WFJ0 A0A2J7Q0R0 B4N542 A0A0M9A7M4 A0A1L8DT79 B4L0N9 A0A2R7WA38 A0A0Q9XLF7 A0A0M4E846 A0A0Q9XNC5 B4LG71 A0A1W4W7A3 A0A0Q9WI37 A0A154PMR7 B4PD71 A0A1W4W6F3 A0A195BFI9 A0A0R1DU99
A0A1W4X5Q9 D6W9I3 A0A2J7Q0N6 A0A1Y1NCD0 A0A2J7Q0N2 A0A1Y1NEM8 A0A067QLX8 E0VG83 A0A182V393 A0A182R9U6 A0A182P132 A0A2C9GS90 Q7PT00 A7URT7 A0A1B6FXJ2 A0A1B6ECJ7 A0A0K8SYD2 A0A0K8SYP3 A0A182Y317 A0A0K8SYD6 A0A1B6ME28 A0A1B6L8H2 A0A1Q3F9M7 A0A1B6G8R1 A0A1Q3F9S7 E2C7G3 A0A224XIS4 A0A182M4V1 A0A023F3C0 A0A182JTG2 A0A1B6KL25 B0X7X1 A0A1B6KIM6 A0A151JBP9 A0A0P4VZS4 A0A182ILN7 A0A084WRP8 A0A2M4AHB6 A0A182WWM0 A0A0V0G4T3 A0A182FGC0 E2AGG4 A0A2M4AHL9 A0A158NRC6 T1IG46 A0A182WHX5 F4WB37 A0A2M4CYM4 A0A1A9V478 A0A195FJJ8 A0A1I8PEI6 W5JA25 A0A088A9B2 A0A2A3EG75 A0A0J7KIS1 A0A151XF13 Q178R9 A0A151IFJ4 A0A1I8MYV6 A0A1I8PEQ1 A0A1I8MYV4 A0A182N9R9 A0A1Y1NBS2 A0A0L7RJK0 A0A336LTB2 A0A0K8V8N7 A0A1A9WLI1 K7JBL0 A0A1B6DL92 A0A0L0BV94 W8B7F6 A0A0C9QA00 A0A0C9R532 A0A232EEZ9 A0A034VTP6 A0A026WFJ0 A0A2J7Q0R0 B4N542 A0A0M9A7M4 A0A1L8DT79 B4L0N9 A0A2R7WA38 A0A0Q9XLF7 A0A0M4E846 A0A0Q9XNC5 B4LG71 A0A1W4W7A3 A0A0Q9WI37 A0A154PMR7 B4PD71 A0A1W4W6F3 A0A195BFI9 A0A0R1DU99
Pubmed
EMBL
BABH01032256
NWSH01004216
PCG65340.1
AGBW02012137
OWR45576.1
KQ458665
+ More
KPJ05602.1 RSAL01000238 RVE43700.1 KQ461198 KPJ06054.1 KQ971312 EEZ98497.1 NEVH01019964 PNF22147.1 GEZM01007124 JAV95359.1 PNF22148.1 GEZM01007123 JAV95360.1 KK853169 KDR10359.1 DS235132 EEB12389.1 APCN01003701 AAAB01008807 EAA04764.5 EDO64729.1 GECZ01014863 JAS54906.1 GEDC01001646 JAS35652.1 GBRD01007524 JAG58297.1 GBRD01007525 JAG58296.1 GBRD01007523 GDHC01021356 JAG58298.1 JAP97272.1 GEBQ01029421 GEBQ01005817 JAT10556.1 JAT34160.1 GEBQ01030074 GEBQ01019968 JAT09903.1 JAT20009.1 GFDL01010764 JAV24281.1 GECZ01010934 JAS58835.1 GFDL01010772 JAV24273.1 GL453369 EFN76150.1 GFTR01008485 JAW07941.1 AXCM01000491 GBBI01003006 JAC15706.1 GEBQ01027834 GEBQ01024072 JAT12143.1 JAT15905.1 DS232468 EDS42190.1 GEBQ01028684 GEBQ01016358 JAT11293.1 JAT23619.1 KQ979122 KYN22546.1 GDKW01000508 JAI56087.1 ATLV01026175 KE525407 KFB52892.1 GGFK01006865 MBW40186.1 GECL01003067 JAP03057.1 GL439310 EFN67497.1 GGFK01006950 MBW40271.1 ADTU01023875 ADTU01023876 ACPB03002682 GL888056 EGI68592.1 GGFL01006123 MBW70301.1 KQ981522 KYN40541.1 ADMH02002070 ETN59614.1 KZ288259 PBC30484.1 LBMM01006983 KMQ90144.1 KQ982205 KYQ58984.1 CH477360 EAT42672.1 KQ977771 KYM99968.1 GEZM01007122 JAV95361.1 KQ414580 KOC71050.1 UFQT01000185 SSX21266.1 GDHF01017062 JAI35252.1 AAZX01008745 GEDC01010890 JAS26408.1 JRES01001407 KNC23164.1 GAMC01009425 JAB97130.1 GBYB01011228 JAG80995.1 GBYB01001956 JAG71723.1 NNAY01005177 OXU16925.1 GAKP01012276 JAC46676.1 KK107238 EZA54688.1 PNF22146.1 CH964101 EDW79481.1 KQ435729 KOX77583.1 GFDF01004415 JAV09669.1 CH933809 EDW18116.1 KK854520 PTY16542.1 KRG05986.1 CP012525 ALC42991.1 KRG05985.1 CH940647 EDW69379.1 KRF84349.1 KQ434984 KZC13149.1 CM000159 EDW92819.1 KQ976504 KYM82935.1 KRK00728.1
KPJ05602.1 RSAL01000238 RVE43700.1 KQ461198 KPJ06054.1 KQ971312 EEZ98497.1 NEVH01019964 PNF22147.1 GEZM01007124 JAV95359.1 PNF22148.1 GEZM01007123 JAV95360.1 KK853169 KDR10359.1 DS235132 EEB12389.1 APCN01003701 AAAB01008807 EAA04764.5 EDO64729.1 GECZ01014863 JAS54906.1 GEDC01001646 JAS35652.1 GBRD01007524 JAG58297.1 GBRD01007525 JAG58296.1 GBRD01007523 GDHC01021356 JAG58298.1 JAP97272.1 GEBQ01029421 GEBQ01005817 JAT10556.1 JAT34160.1 GEBQ01030074 GEBQ01019968 JAT09903.1 JAT20009.1 GFDL01010764 JAV24281.1 GECZ01010934 JAS58835.1 GFDL01010772 JAV24273.1 GL453369 EFN76150.1 GFTR01008485 JAW07941.1 AXCM01000491 GBBI01003006 JAC15706.1 GEBQ01027834 GEBQ01024072 JAT12143.1 JAT15905.1 DS232468 EDS42190.1 GEBQ01028684 GEBQ01016358 JAT11293.1 JAT23619.1 KQ979122 KYN22546.1 GDKW01000508 JAI56087.1 ATLV01026175 KE525407 KFB52892.1 GGFK01006865 MBW40186.1 GECL01003067 JAP03057.1 GL439310 EFN67497.1 GGFK01006950 MBW40271.1 ADTU01023875 ADTU01023876 ACPB03002682 GL888056 EGI68592.1 GGFL01006123 MBW70301.1 KQ981522 KYN40541.1 ADMH02002070 ETN59614.1 KZ288259 PBC30484.1 LBMM01006983 KMQ90144.1 KQ982205 KYQ58984.1 CH477360 EAT42672.1 KQ977771 KYM99968.1 GEZM01007122 JAV95361.1 KQ414580 KOC71050.1 UFQT01000185 SSX21266.1 GDHF01017062 JAI35252.1 AAZX01008745 GEDC01010890 JAS26408.1 JRES01001407 KNC23164.1 GAMC01009425 JAB97130.1 GBYB01011228 JAG80995.1 GBYB01001956 JAG71723.1 NNAY01005177 OXU16925.1 GAKP01012276 JAC46676.1 KK107238 EZA54688.1 PNF22146.1 CH964101 EDW79481.1 KQ435729 KOX77583.1 GFDF01004415 JAV09669.1 CH933809 EDW18116.1 KK854520 PTY16542.1 KRG05986.1 CP012525 ALC42991.1 KRG05985.1 CH940647 EDW69379.1 KRF84349.1 KQ434984 KZC13149.1 CM000159 EDW92819.1 KQ976504 KYM82935.1 KRK00728.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
+ More
UP000192223 UP000007266 UP000235965 UP000027135 UP000009046 UP000075903 UP000075900 UP000075885 UP000075840 UP000007062 UP000076408 UP000008237 UP000075883 UP000075881 UP000002320 UP000078492 UP000075880 UP000030765 UP000076407 UP000069272 UP000000311 UP000005205 UP000015103 UP000075920 UP000007755 UP000078200 UP000078541 UP000095300 UP000000673 UP000005203 UP000242457 UP000036403 UP000075809 UP000008820 UP000078542 UP000095301 UP000075884 UP000053825 UP000091820 UP000002358 UP000037069 UP000215335 UP000053097 UP000007798 UP000053105 UP000009192 UP000092553 UP000008792 UP000192221 UP000076502 UP000002282 UP000078540
UP000192223 UP000007266 UP000235965 UP000027135 UP000009046 UP000075903 UP000075900 UP000075885 UP000075840 UP000007062 UP000076408 UP000008237 UP000075883 UP000075881 UP000002320 UP000078492 UP000075880 UP000030765 UP000076407 UP000069272 UP000000311 UP000005205 UP000015103 UP000075920 UP000007755 UP000078200 UP000078541 UP000095300 UP000000673 UP000005203 UP000242457 UP000036403 UP000075809 UP000008820 UP000078542 UP000095301 UP000075884 UP000053825 UP000091820 UP000002358 UP000037069 UP000215335 UP000053097 UP000007798 UP000053105 UP000009192 UP000092553 UP000008792 UP000192221 UP000076502 UP000002282 UP000078540
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JSR6
A0A2A4J1M4
A0A212EVR6
A0A0N1PGC6
A0A3S2P7H9
A0A194QKY1
+ More
A0A1W4X5Q9 D6W9I3 A0A2J7Q0N6 A0A1Y1NCD0 A0A2J7Q0N2 A0A1Y1NEM8 A0A067QLX8 E0VG83 A0A182V393 A0A182R9U6 A0A182P132 A0A2C9GS90 Q7PT00 A7URT7 A0A1B6FXJ2 A0A1B6ECJ7 A0A0K8SYD2 A0A0K8SYP3 A0A182Y317 A0A0K8SYD6 A0A1B6ME28 A0A1B6L8H2 A0A1Q3F9M7 A0A1B6G8R1 A0A1Q3F9S7 E2C7G3 A0A224XIS4 A0A182M4V1 A0A023F3C0 A0A182JTG2 A0A1B6KL25 B0X7X1 A0A1B6KIM6 A0A151JBP9 A0A0P4VZS4 A0A182ILN7 A0A084WRP8 A0A2M4AHB6 A0A182WWM0 A0A0V0G4T3 A0A182FGC0 E2AGG4 A0A2M4AHL9 A0A158NRC6 T1IG46 A0A182WHX5 F4WB37 A0A2M4CYM4 A0A1A9V478 A0A195FJJ8 A0A1I8PEI6 W5JA25 A0A088A9B2 A0A2A3EG75 A0A0J7KIS1 A0A151XF13 Q178R9 A0A151IFJ4 A0A1I8MYV6 A0A1I8PEQ1 A0A1I8MYV4 A0A182N9R9 A0A1Y1NBS2 A0A0L7RJK0 A0A336LTB2 A0A0K8V8N7 A0A1A9WLI1 K7JBL0 A0A1B6DL92 A0A0L0BV94 W8B7F6 A0A0C9QA00 A0A0C9R532 A0A232EEZ9 A0A034VTP6 A0A026WFJ0 A0A2J7Q0R0 B4N542 A0A0M9A7M4 A0A1L8DT79 B4L0N9 A0A2R7WA38 A0A0Q9XLF7 A0A0M4E846 A0A0Q9XNC5 B4LG71 A0A1W4W7A3 A0A0Q9WI37 A0A154PMR7 B4PD71 A0A1W4W6F3 A0A195BFI9 A0A0R1DU99
A0A1W4X5Q9 D6W9I3 A0A2J7Q0N6 A0A1Y1NCD0 A0A2J7Q0N2 A0A1Y1NEM8 A0A067QLX8 E0VG83 A0A182V393 A0A182R9U6 A0A182P132 A0A2C9GS90 Q7PT00 A7URT7 A0A1B6FXJ2 A0A1B6ECJ7 A0A0K8SYD2 A0A0K8SYP3 A0A182Y317 A0A0K8SYD6 A0A1B6ME28 A0A1B6L8H2 A0A1Q3F9M7 A0A1B6G8R1 A0A1Q3F9S7 E2C7G3 A0A224XIS4 A0A182M4V1 A0A023F3C0 A0A182JTG2 A0A1B6KL25 B0X7X1 A0A1B6KIM6 A0A151JBP9 A0A0P4VZS4 A0A182ILN7 A0A084WRP8 A0A2M4AHB6 A0A182WWM0 A0A0V0G4T3 A0A182FGC0 E2AGG4 A0A2M4AHL9 A0A158NRC6 T1IG46 A0A182WHX5 F4WB37 A0A2M4CYM4 A0A1A9V478 A0A195FJJ8 A0A1I8PEI6 W5JA25 A0A088A9B2 A0A2A3EG75 A0A0J7KIS1 A0A151XF13 Q178R9 A0A151IFJ4 A0A1I8MYV6 A0A1I8PEQ1 A0A1I8MYV4 A0A182N9R9 A0A1Y1NBS2 A0A0L7RJK0 A0A336LTB2 A0A0K8V8N7 A0A1A9WLI1 K7JBL0 A0A1B6DL92 A0A0L0BV94 W8B7F6 A0A0C9QA00 A0A0C9R532 A0A232EEZ9 A0A034VTP6 A0A026WFJ0 A0A2J7Q0R0 B4N542 A0A0M9A7M4 A0A1L8DT79 B4L0N9 A0A2R7WA38 A0A0Q9XLF7 A0A0M4E846 A0A0Q9XNC5 B4LG71 A0A1W4W7A3 A0A0Q9WI37 A0A154PMR7 B4PD71 A0A1W4W6F3 A0A195BFI9 A0A0R1DU99
PDB
1D5F
E-value=4.88996e-78,
Score=745
Ontologies
GO
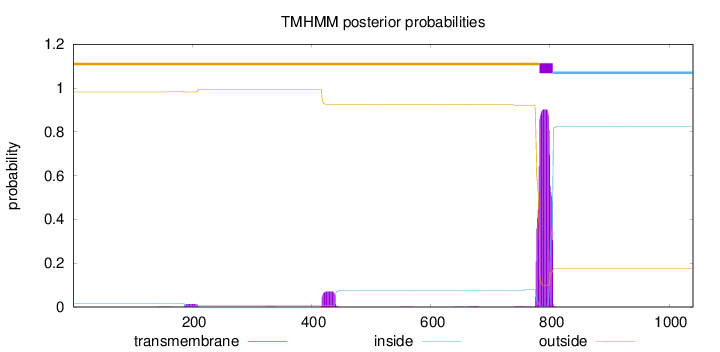
Topology
Length:
1040
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.70462
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.01755
outside
1 - 782
TMhelix
783 - 805
inside
806 - 1040
Population Genetic Test Statistics
Pi
251.710433
Theta
171.263096
Tajima's D
1.492414
CLR
0.350527
CSRT
0.784210789460527
Interpretation
Uncertain
