Gene
KWMTBOMO05016 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012524
Annotation
PREDICTED:_activating_signal_cointegrator_1_complex_subunit_2_homolog_isoform_X4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.167
Sequence
CDS
ATGGCCACCGGCCCGAAGATAGTACACAAGCAGTTCAACTCGCCCATCGGGTTGTACTCGCAGCAGAATATCAAGGAAACGCTCAACAAACACCTGAAAAACCTCGACAACGGATCTGTCGGAATCGACTTCAACAACCCGACAACGGATAAGCCAGCGAACTTAGCTAATTCCGCCGTCCTAAGAATGTTAGAAGAGGAAGAGCGTAACCGCAAAGGATACAGTCAGAAGAAAGTGGTGTGGCCTCCAGTACCCGAAACGAACGGATATCATAATCCACAACAGCAGAGTCCCGTGTACGACAACAACGCGCAACACTCCCCGTCTGCTCAGCCTCCCTATCAAAAGCAACCTCAATATCAACCCGAGCCCCAATATCAAGCTCAGCCTCCGCACTTCCAACCCCAATACCAACAGCCACAGTCCCAGCAGCCTCAGTATCCCCAGTCTCAGCCAGGGTTCTGTGAGGGACGTCGTCAAGAGCAAACTGGCCTGAGTAAGCTGATTCCCGTTGGGCCGGGAGTGACTGCGTCACCTTTGTCTCCCTCGCAGTACCGTGTGGGTCCGGCGTCCCCGGGCATCGCACCGCAACCCTTCAGGCCACAGCCAAATCGCTGGGCCCCCGTACCGGCCCCCCTCTCCCCTCAGACACCCGGATCGCAGCCGAGCTACACGCAGCAGATTTCCCCTCAATCTCAGTACTCATCGCAGTCTCCTTATTCGGAGCCCGCGTACCAGACCACACAGAGGCACTTCGAACCACCTCCATCAACCATCACTCTCCGTCCGCAACCTCCGGTTCACCAGAAACCATCCCCGGTGTTCGCTAGTCAACCGGCCGCTGCGTCATTTAAAGGAGGCGTCAACATGCGTGGAGATCAGAAGTGGCCGCCTCAATCGGTGAAGGAGGCTGTGGCTGCAGAAAACGAAGCCCGCCTCAAACTAGCCAAGGGTCCCGCCTGCAGACCGCGCAAGGTTAAAAAGGACTACAGCTCGTTCTTCGCGAAGCATGCCCTGAGCCACAATTACCCCGGCTACCGCGCGCCACCAGGCACCCAGCACTTCGAAGACTTCTCGGGCCGATCCTCATACTAA
Protein
MATGPKIVHKQFNSPIGLYSQQNIKETLNKHLKNLDNGSVGIDFNNPTTDKPANLANSAVLRMLEEEERNRKGYSQKKVVWPPVPETNGYHNPQQQSPVYDNNAQHSPSAQPPYQKQPQYQPEPQYQAQPPHFQPQYQQPQSQQPQYPQSQPGFCEGRRQEQTGLSKLIPVGPGVTASPLSPSQYRVGPASPGIAPQPFRPQPNRWAPVPAPLSPQTPGSQPSYTQQISPQSQYSSQSPYSEPAYQTTQRHFEPPPSTITLRPQPPVHQKPSPVFASQPAAASFKGGVNMRGDQKWPPQSVKEAVAAENEARLKLAKGPACRPRKVKKDYSSFFAKHALSHNYPGYRAPPGTQHFEDFSGRSSY
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF15936 DUF4749
ProteinModelPortal
Ontologies
GO
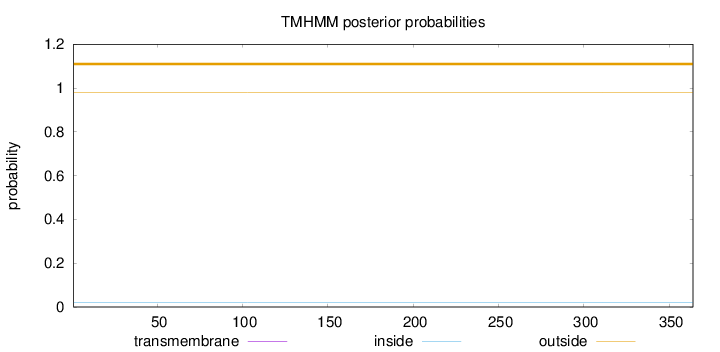
Topology
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00035
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01969
outside
1 - 364
Population Genetic Test Statistics
Pi
244.183196
Theta
188.274377
Tajima's D
1.203084
CLR
0.289881
CSRT
0.713764311784411
Interpretation
Uncertain
