Pre Gene Modal
BGIBMGA008117
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_13C_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 13
Location in the cell
Nuclear Reliability : 2.172
Sequence
CDS
ATGCAGTATGCGGTTCATCATCACACGTTCATCGATTTAGATATCGACATTGCCTCCTCTTATATTATTGTACCGCAGACGGGAAAATACCAAGGAAAAGAGTCGTGTGTTGTCGTGAAACTTGGTGCTATAAACGTCAAGTCCGAACCTCGCTCCCAAGCCCTCGACGTACACAAATTGTATAAAGAAGGATTGGCCGACGAAGATATTCTGCGTGAGATAACCCAGCACAGCTACGATAAGATGGCACTGAAACTAACCGAAATGCAGATCGTTATAGCAACGTCGTCGGAAGATTGGCAGTCGGCTATAGCTTCGAACGAAGCGACGTCCCTGCACTTGCTGCAGCCAACGAACTTGGTCATACAAATACACAAATGTCTGATCACGGACGATCCGAGAATGCCGAAGGTCAAAGTCCGGGGTGAACTCGAGAAAATCGCTATAAGCGCCTCCGAAGACCGATTGCTGACACTTTGTGAAATATTAGTCAGTATGCCGTTACCTAATTCAGAAGAATCGGCTCAGTTGAAGCCAAGCGACTCGAAAGGATCAAGTTTGTCCTTACTGAAGTATTTAGATCCGGCTGAAAAACAAAAGAGAGACACTGCTCGCTCAAAGGAGCGGAAGAGTGACGAGCAAACTATACAGTTAACTGAAGTTGAAGCCTTCTTTATAATGAAAGGTAACGTAACGTGA
Protein
MQYAVHHHTFIDLDIDIASSYIIVPQTGKYQGKESCVVVKLGAINVKSEPRSQALDVHKLYKEGLADEDILREITQHSYDKMALKLTEMQIVIATSSEDWQSAIASNEATSLHLLQPTNLVIQIHKCLITDDPRMPKVKVRGELEKIAISASEDRLLTLCEILVSMPLPNSEESAQLKPSDSKGSSLSLLKYLDPAEKQKRDTARSKERKSDEQTIQLTEVEAFFIMKGNVT
Summary
Description
Plays a role in the survival of neurons by maintaining protein homeostasis in the central nervous system. May function as part of a lysosomal degradation pathway.
Similarity
Belongs to the VPS13 family.
Keywords
Complete proteome
Endosome
Membrane
Protein transport
Reference proteome
Transport
Feature
chain Vacuolar protein sorting-associated protein 13
Uniprot
H9JF20
A0A3S2LSU5
A0A2A4JAS4
A0A0N0PAP2
A0A194QM57
A0A1E1VZK4
+ More
A0A1E1WNG3 A0A212EH99 A0A067RTP5 A0A182MPK1 A0A182SUD4 A0A182WB65 A0A182RA29 A0A182NLZ6 A0A182PLD4 A0A182IQ42 A0A182QNC8 A0A182LB90 Q5TSU0 A0A182HPA7 A0A182VBQ3 A0A182TV09 A0A182X4A3 A0A182JW81 A0A084WJG5 A0A1Q3FGM4 A0A1Q3FGD3 A0A182G3T1 B0WUG9 A0A182YFP8 A0A336L615 Q16SR9 A0A2P8Y000 A0A1S4E8Q6 A0A2H1W0S9 A0A2M3ZZF8 W4VRJ2 D6WCW1 W5J689 A0A2M4DRU1 A0A0P6JRN7 A0A1J1ITZ1 A0A2M4CVW7 A0A182FL84 V5GY02 N6U474 U4ULV9 A0A1Y1M0H6 A0A1L8DVL7 A0A232F955 J9K7I7 K7IXV4 A0A2H8TPG9 A0A2H8TP76 A0A1B6CUA0 E9J4Z2 T1J4H6 A0A0J7KFM8 E1ZV01 A0A3L8DLF8 A0A026WHS8 V9IL78 A0A0P4WPI9 A0A0P4W8G3 A0A154PLH2 A0A0P4WAQ3 A0A0P4W473 A0A151JQF4 E2B476 A0A1B0DB55 A0A151JY96 A0A195CYF9 A0A151X236 A0A3R7PJS0 A0A2A3EL42 A0A087ZQS4 A0A310S5Y6 B4NS47 A0A0N0BCF1 B3MHR1 A0A0P8XPG8 A0A0V0GCP3 A0A0L7RCF4 A0A1I8PU10 A0A1B6LDL1 B4MIP9 A0A1B6KWT9 A0A224XG50 A0A1I8NHV4 T1P7V0 A0A0T6BDC0 A0A0J9R7H7 B4P1S0 A0A023EZM1 A1Z713 E0VJH2
A0A1E1WNG3 A0A212EH99 A0A067RTP5 A0A182MPK1 A0A182SUD4 A0A182WB65 A0A182RA29 A0A182NLZ6 A0A182PLD4 A0A182IQ42 A0A182QNC8 A0A182LB90 Q5TSU0 A0A182HPA7 A0A182VBQ3 A0A182TV09 A0A182X4A3 A0A182JW81 A0A084WJG5 A0A1Q3FGM4 A0A1Q3FGD3 A0A182G3T1 B0WUG9 A0A182YFP8 A0A336L615 Q16SR9 A0A2P8Y000 A0A1S4E8Q6 A0A2H1W0S9 A0A2M3ZZF8 W4VRJ2 D6WCW1 W5J689 A0A2M4DRU1 A0A0P6JRN7 A0A1J1ITZ1 A0A2M4CVW7 A0A182FL84 V5GY02 N6U474 U4ULV9 A0A1Y1M0H6 A0A1L8DVL7 A0A232F955 J9K7I7 K7IXV4 A0A2H8TPG9 A0A2H8TP76 A0A1B6CUA0 E9J4Z2 T1J4H6 A0A0J7KFM8 E1ZV01 A0A3L8DLF8 A0A026WHS8 V9IL78 A0A0P4WPI9 A0A0P4W8G3 A0A154PLH2 A0A0P4WAQ3 A0A0P4W473 A0A151JQF4 E2B476 A0A1B0DB55 A0A151JY96 A0A195CYF9 A0A151X236 A0A3R7PJS0 A0A2A3EL42 A0A087ZQS4 A0A310S5Y6 B4NS47 A0A0N0BCF1 B3MHR1 A0A0P8XPG8 A0A0V0GCP3 A0A0L7RCF4 A0A1I8PU10 A0A1B6LDL1 B4MIP9 A0A1B6KWT9 A0A224XG50 A0A1I8NHV4 T1P7V0 A0A0T6BDC0 A0A0J9R7H7 B4P1S0 A0A023EZM1 A1Z713 E0VJH2
Pubmed
19121390
26354079
22118469
24845553
20966253
12364791
+ More
24438588 26483478 25244985 17510324 29403074 18362917 19820115 20920257 23761445 26999592 23537049 28004739 28648823 20075255 21282665 20798317 30249741 24508170 17994087 18057021 25315136 22936249 17550304 25474469 10731132 12537572 28107480 20566863
24438588 26483478 25244985 17510324 29403074 18362917 19820115 20920257 23761445 26999592 23537049 28004739 28648823 20075255 21282665 20798317 30249741 24508170 17994087 18057021 25315136 22936249 17550304 25474469 10731132 12537572 28107480 20566863
EMBL
BABH01033962
BABH01033963
BABH01033964
BABH01033965
BABH01033966
BABH01033967
+ More
RSAL01000238 RVE43697.1 NWSH01002177 PCG68959.1 KQ458665 KPJ05606.1 KQ461198 KPJ06050.1 GDQN01010879 JAT80175.1 GDQN01002587 JAT88467.1 AGBW02014919 OWR40856.1 KK852470 KDR23219.1 AXCM01006669 AXCN02000957 AAAB01008900 EAL40459.3 APCN01002939 ATLV01024026 ATLV01024027 KE525348 KFB50359.1 GFDL01008393 JAV26652.1 GFDL01008441 JAV26604.1 JXUM01007029 JXUM01007030 KQ560229 KXJ83724.1 DS232105 EDS34951.1 UFQS01001985 UFQT01001985 SSX12864.1 SSX32306.1 CH477667 EAT37516.1 PYGN01001102 PSN37570.1 ODYU01005601 SOQ46613.1 GGFK01000558 MBW33879.1 GANO01002385 JAB57486.1 KQ971311 EEZ98831.2 ADMH02002077 ETN59516.1 GGFL01016096 MBW80274.1 GDUN01001099 JAN94820.1 CVRI01000059 CRL03731.1 GGFL01005306 MBW69484.1 GALX01003208 JAB65258.1 APGK01040378 KB740979 ENN76405.1 KB632379 ERL94107.1 GEZM01043725 JAV78881.1 GFDF01003625 JAV10459.1 NNAY01000686 OXU26973.1 ABLF02021408 ABLF02021409 GFXV01003333 MBW15138.1 GFXV01003995 MBW15800.1 GEDC01020311 JAS16987.1 GL768121 EFZ12134.1 JH431845 LBMM01008239 KMQ89031.1 GL434335 EFN74999.1 QOIP01000007 RLU20668.1 KK107238 EZA54619.1 JR052898 AEY61810.1 GDRN01073504 JAI63382.1 GDRN01073508 JAI63379.1 KQ434960 KZC12683.1 GDRN01073505 JAI63381.1 GDRN01073507 JAI63380.1 KQ978684 KYN29306.1 GL445515 EFN89493.1 AJVK01000756 AJVK01000757 AJVK01000758 AJVK01000759 AJVK01000760 KQ981561 KYN39826.1 KQ977171 KYN05179.1 KQ982584 KYQ54431.1 QCYY01002627 ROT68842.1 KZ288226 PBC31869.1 KQ769179 OAD52957.1 CH981877 EDX15425.1 KQ435911 KOX68931.1 CH902619 EDV36898.1 KPU76493.1 KPU76494.1 GECL01000527 JAP05597.1 KQ414616 KOC68416.1 GEBQ01018189 JAT21788.1 CH963719 EDW71988.1 GEBQ01024054 JAT15923.1 GFTR01008996 JAW07430.1 KA644701 AFP59330.1 LJIG01001609 KRT85331.1 CM002911 KMY92020.1 KMY92021.1 CM000157 EDW89206.2 KRJ98276.1 GBBI01004017 JAC14695.1 AE013599 DS235222 EEB13528.1
RSAL01000238 RVE43697.1 NWSH01002177 PCG68959.1 KQ458665 KPJ05606.1 KQ461198 KPJ06050.1 GDQN01010879 JAT80175.1 GDQN01002587 JAT88467.1 AGBW02014919 OWR40856.1 KK852470 KDR23219.1 AXCM01006669 AXCN02000957 AAAB01008900 EAL40459.3 APCN01002939 ATLV01024026 ATLV01024027 KE525348 KFB50359.1 GFDL01008393 JAV26652.1 GFDL01008441 JAV26604.1 JXUM01007029 JXUM01007030 KQ560229 KXJ83724.1 DS232105 EDS34951.1 UFQS01001985 UFQT01001985 SSX12864.1 SSX32306.1 CH477667 EAT37516.1 PYGN01001102 PSN37570.1 ODYU01005601 SOQ46613.1 GGFK01000558 MBW33879.1 GANO01002385 JAB57486.1 KQ971311 EEZ98831.2 ADMH02002077 ETN59516.1 GGFL01016096 MBW80274.1 GDUN01001099 JAN94820.1 CVRI01000059 CRL03731.1 GGFL01005306 MBW69484.1 GALX01003208 JAB65258.1 APGK01040378 KB740979 ENN76405.1 KB632379 ERL94107.1 GEZM01043725 JAV78881.1 GFDF01003625 JAV10459.1 NNAY01000686 OXU26973.1 ABLF02021408 ABLF02021409 GFXV01003333 MBW15138.1 GFXV01003995 MBW15800.1 GEDC01020311 JAS16987.1 GL768121 EFZ12134.1 JH431845 LBMM01008239 KMQ89031.1 GL434335 EFN74999.1 QOIP01000007 RLU20668.1 KK107238 EZA54619.1 JR052898 AEY61810.1 GDRN01073504 JAI63382.1 GDRN01073508 JAI63379.1 KQ434960 KZC12683.1 GDRN01073505 JAI63381.1 GDRN01073507 JAI63380.1 KQ978684 KYN29306.1 GL445515 EFN89493.1 AJVK01000756 AJVK01000757 AJVK01000758 AJVK01000759 AJVK01000760 KQ981561 KYN39826.1 KQ977171 KYN05179.1 KQ982584 KYQ54431.1 QCYY01002627 ROT68842.1 KZ288226 PBC31869.1 KQ769179 OAD52957.1 CH981877 EDX15425.1 KQ435911 KOX68931.1 CH902619 EDV36898.1 KPU76493.1 KPU76494.1 GECL01000527 JAP05597.1 KQ414616 KOC68416.1 GEBQ01018189 JAT21788.1 CH963719 EDW71988.1 GEBQ01024054 JAT15923.1 GFTR01008996 JAW07430.1 KA644701 AFP59330.1 LJIG01001609 KRT85331.1 CM002911 KMY92020.1 KMY92021.1 CM000157 EDW89206.2 KRJ98276.1 GBBI01004017 JAC14695.1 AE013599 DS235222 EEB13528.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000027135 UP000075883 UP000075901 UP000075920 UP000075900 UP000075884 UP000075885 UP000075880 UP000075886 UP000075882 UP000007062 UP000075840 UP000075903 UP000075902 UP000076407 UP000075881 UP000030765 UP000069940 UP000249989 UP000002320 UP000076408 UP000008820 UP000245037 UP000079169 UP000007266 UP000000673 UP000183832 UP000069272 UP000019118 UP000030742 UP000215335 UP000007819 UP000002358 UP000036403 UP000000311 UP000279307 UP000053097 UP000076502 UP000078492 UP000008237 UP000092462 UP000078541 UP000078542 UP000075809 UP000283509 UP000242457 UP000005203 UP000000304 UP000053105 UP000007801 UP000053825 UP000095300 UP000007798 UP000095301 UP000002282 UP000000803 UP000009046
UP000027135 UP000075883 UP000075901 UP000075920 UP000075900 UP000075884 UP000075885 UP000075880 UP000075886 UP000075882 UP000007062 UP000075840 UP000075903 UP000075902 UP000076407 UP000075881 UP000030765 UP000069940 UP000249989 UP000002320 UP000076408 UP000008820 UP000245037 UP000079169 UP000007266 UP000000673 UP000183832 UP000069272 UP000019118 UP000030742 UP000215335 UP000007819 UP000002358 UP000036403 UP000000311 UP000279307 UP000053097 UP000076502 UP000078492 UP000008237 UP000092462 UP000078541 UP000078542 UP000075809 UP000283509 UP000242457 UP000005203 UP000000304 UP000053105 UP000007801 UP000053825 UP000095300 UP000007798 UP000095301 UP000002282 UP000000803 UP000009046
Pfam
Interpro
ProteinModelPortal
H9JF20
A0A3S2LSU5
A0A2A4JAS4
A0A0N0PAP2
A0A194QM57
A0A1E1VZK4
+ More
A0A1E1WNG3 A0A212EH99 A0A067RTP5 A0A182MPK1 A0A182SUD4 A0A182WB65 A0A182RA29 A0A182NLZ6 A0A182PLD4 A0A182IQ42 A0A182QNC8 A0A182LB90 Q5TSU0 A0A182HPA7 A0A182VBQ3 A0A182TV09 A0A182X4A3 A0A182JW81 A0A084WJG5 A0A1Q3FGM4 A0A1Q3FGD3 A0A182G3T1 B0WUG9 A0A182YFP8 A0A336L615 Q16SR9 A0A2P8Y000 A0A1S4E8Q6 A0A2H1W0S9 A0A2M3ZZF8 W4VRJ2 D6WCW1 W5J689 A0A2M4DRU1 A0A0P6JRN7 A0A1J1ITZ1 A0A2M4CVW7 A0A182FL84 V5GY02 N6U474 U4ULV9 A0A1Y1M0H6 A0A1L8DVL7 A0A232F955 J9K7I7 K7IXV4 A0A2H8TPG9 A0A2H8TP76 A0A1B6CUA0 E9J4Z2 T1J4H6 A0A0J7KFM8 E1ZV01 A0A3L8DLF8 A0A026WHS8 V9IL78 A0A0P4WPI9 A0A0P4W8G3 A0A154PLH2 A0A0P4WAQ3 A0A0P4W473 A0A151JQF4 E2B476 A0A1B0DB55 A0A151JY96 A0A195CYF9 A0A151X236 A0A3R7PJS0 A0A2A3EL42 A0A087ZQS4 A0A310S5Y6 B4NS47 A0A0N0BCF1 B3MHR1 A0A0P8XPG8 A0A0V0GCP3 A0A0L7RCF4 A0A1I8PU10 A0A1B6LDL1 B4MIP9 A0A1B6KWT9 A0A224XG50 A0A1I8NHV4 T1P7V0 A0A0T6BDC0 A0A0J9R7H7 B4P1S0 A0A023EZM1 A1Z713 E0VJH2
A0A1E1WNG3 A0A212EH99 A0A067RTP5 A0A182MPK1 A0A182SUD4 A0A182WB65 A0A182RA29 A0A182NLZ6 A0A182PLD4 A0A182IQ42 A0A182QNC8 A0A182LB90 Q5TSU0 A0A182HPA7 A0A182VBQ3 A0A182TV09 A0A182X4A3 A0A182JW81 A0A084WJG5 A0A1Q3FGM4 A0A1Q3FGD3 A0A182G3T1 B0WUG9 A0A182YFP8 A0A336L615 Q16SR9 A0A2P8Y000 A0A1S4E8Q6 A0A2H1W0S9 A0A2M3ZZF8 W4VRJ2 D6WCW1 W5J689 A0A2M4DRU1 A0A0P6JRN7 A0A1J1ITZ1 A0A2M4CVW7 A0A182FL84 V5GY02 N6U474 U4ULV9 A0A1Y1M0H6 A0A1L8DVL7 A0A232F955 J9K7I7 K7IXV4 A0A2H8TPG9 A0A2H8TP76 A0A1B6CUA0 E9J4Z2 T1J4H6 A0A0J7KFM8 E1ZV01 A0A3L8DLF8 A0A026WHS8 V9IL78 A0A0P4WPI9 A0A0P4W8G3 A0A154PLH2 A0A0P4WAQ3 A0A0P4W473 A0A151JQF4 E2B476 A0A1B0DB55 A0A151JY96 A0A195CYF9 A0A151X236 A0A3R7PJS0 A0A2A3EL42 A0A087ZQS4 A0A310S5Y6 B4NS47 A0A0N0BCF1 B3MHR1 A0A0P8XPG8 A0A0V0GCP3 A0A0L7RCF4 A0A1I8PU10 A0A1B6LDL1 B4MIP9 A0A1B6KWT9 A0A224XG50 A0A1I8NHV4 T1P7V0 A0A0T6BDC0 A0A0J9R7H7 B4P1S0 A0A023EZM1 A1Z713 E0VJH2
Ontologies
PANTHER
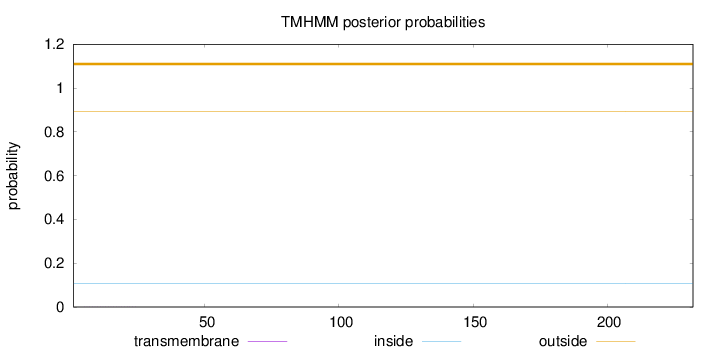
Topology
Subcellular location
Late endosome membrane
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00408999999999999
Exp number, first 60 AAs:
0.00396
Total prob of N-in:
0.10690
outside
1 - 232
Population Genetic Test Statistics
Pi
226.79769
Theta
188.799998
Tajima's D
0.570197
CLR
0.059536
CSRT
0.532923353832308
Interpretation
Uncertain
