Gene
KWMTBOMO05009
Pre Gene Modal
BGIBMGA014408
Annotation
polyprotein_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.182 Nuclear Reliability : 1.686
Sequence
CDS
ATGGAAGGTATCACAGGAAAGCTTCAGATCGGCCTCCTAGATGGAAAAAACTGGGACACCTGGAAATACAAAGCCTTATGCTTACTACGCACCGTACCGTCGGCCCTAGAAGTAGTTGAAGGATCGTTCCAGGAGCCTACTCGTCCACCTACCGGATGCGGCGATGCTGAATTGGCGGCTTATCAACAACAAAAAGATAAGTTTGATAAAGCTGATGCCAGCGCTATCCTAATACTTACCACCAATATGCAAGAAGACACACTTCGAAAAGTTATGCGGTTCACTAAAGTACGTGATGTCTGGTTAGAGCTCCACAGGCTGTTCGACGGAACAAATGCGGACAAAACCTACAACCTGTGTATGAGTTTCTTCAGCTTTAAAAGGGATCAAGCAGACGACATAGCCACTCACATGTCAAAATTAAAGAACATTTGGACTGAACTGAATCAAGAACTCAAGAAGGACAGTAACCAAGAGCTACCCGAACTGCTACTTGTTTGTAAGATACTGGACACGCTTGATGATTCATACTTCAGCTTCAGAAGTAGTTGGCTCCTACTGCCCAAATCCAACCGTACTGTAGAAAACCTAACTAGTCATCTCTGCGCATTTGAAAGGGCACTGCAAGGTATCAACGTACCTTCGCATCATTCGGAAGCCCTGGTTTCGAATACGGAAAAGAAGAAGGACAAGAAGCTTAAATGCAACTACTGCCTAGGTATTGGCCACAGAGTACGCAACTGTCAAAAATGGATCCGTGACGGTAGACCTCCTAAGAGCCAAAATCCGACCCAGACAGCAACGCCTTCTACTTCAAAGACCGCTAATTTGTTACTCATGTCTTTAGAAGACAACGAAATCTTCACAGTTTCACGGGACAACGAGAACTGGTACATCGATAATGGTGCAACAAGCCACGTCACCAACAGAGCTGATTTTTTCCAAACCTACGAACATTTCAATGAGAACTACACTGTTACAACGGCCGCTGGTAATGAAGTCCGTGCCAAAGGCAAAGGAACAGTACAGCTCAGAGGATATGTTAACGGGAAACAAATCGACATCAGCCTGCAAGACGTCTGGTACGTTCCGGCCATAAGCAAGAACCTCTTCTCCATACTCGCACTTCACGACCGAGTCCCGAATAGCACTTTTTCATCCAGGACAACTGAATGCAACGTCATTGTCAACGGGAAAGTATGCTTAATAGGGAAGAGACAAAAACAAGGTGGCTTATATCGACTGGAAGTAAAACCCGTACTGCCAGAAAAACCACCCGAAGTCTTTGCAACTACAGCAACGATGCAACTGTACCACGAGAGACTCGCTCACCAAAATAAAAGACACGCAAGAGCGACGATCCAACGCGAACTAGGTCTAAACCTGCCGATAAATAAAGAGACCTGTGAAGGCTGCATCTTTAGAAAGGCGCATCGACTTAAATTCGGTACAAGAACCCGAGCTACCGCGCCCGGAGAAATTGTACACACCGACGTTTGCGGACCGTTTCAATCTAGCTTCTCAAGCTATAAGTACTACGTTCTGTTCAAAGACGACTTTACTGGCTATCGTATGGTGTACTTCATACGCAATAAGTCGGACACACCATAA
Protein
MEGITGKLQIGLLDGKNWDTWKYKALCLLRTVPSALEVVEGSFQEPTRPPTGCGDAELAAYQQQKDKFDKADASAILILTTNMQEDTLRKVMRFTKVRDVWLELHRLFDGTNADKTYNLCMSFFSFKRDQADDIATHMSKLKNIWTELNQELKKDSNQELPELLLVCKILDTLDDSYFSFRSSWLLLPKSNRTVENLTSHLCAFERALQGINVPSHHSEALVSNTEKKKDKKLKCNYCLGIGHRVRNCQKWIRDGRPPKSQNPTQTATPSTSKTANLLLMSLEDNEIFTVSRDNENWYIDNGATSHVTNRADFFQTYEHFNENYTVTTAAGNEVRAKGKGTVQLRGYVNGKQIDISLQDVWYVPAISKNLFSILALHDRVPNSTFSSRTTECNVIVNGKVCLIGKRQKQGGLYRLEVKPVLPEKPPEVFATTATMQLYHERLAHQNKRHARATIQRELGLNLPINKETCEGCIFRKAHRLKFGTRTRATAPGEIVHTDVCGPFQSSFSSYKYYVLFKDDFTGYRMVYFIRNKSDTP
Summary
Uniprot
B3Y003
A0A1E1WKD7
A0A087T2K3
A0A087SYT1
A0A2J7PN24
A0A1Y1KNM9
+ More
A0A1Y1K3Q7 A0A0V1M1K0 A0A0V0Z7R7 A0A1Y1LRT7 A0A0V1B5K0 A0A0V1B5A2 A0A0V1L945 A0A0J7K744 A0A0V0TK25 A0A0V0TK20 A0A0V1NHV8 J9LCG1 A0A139WFV5 A0A0V1HEX2 A0A2S2P896 A0A1Y1MYL8 A0A1Y1K2V0 A0A1Y1N6B6 X1WMG8 A0A1Y1MYL6 A0A1Y1NHZ0 A0A151ITN2 A0A0V0WE00 A0A0V0WA51 A0A0J7K9V7 A0A0V0W669 A0A1Y1KZM0 A0A0V1G2I5 X1XQX4 A0A2J7QWT7 A0A085N6E0 A0A0V1KG52 A0A0J7K763 X1XSG1 A0A0V1EYB7 A0A0J7N8C0 K7JBX1 K7J7F1 A0A0V1DTZ3 A0A0P5PQ39 K7JXE5 A0A1Y1NEM6 A0A1Y1L0K1 A0A0J7K2P0 A0A0J7K717 A0A0A9YZ05 A0A0A9WFQ0 K7JBT3 A0A085MSN1 A0A0J7K689 A0A1Y1LCN3 A0A0L0CMY6 K7JAW3 A0A0N5E2R3 A0A2A4JFW7 A0A0J7K651 X1WT66 A0A0J7KDE2 A0A1Y1KR09 A0A151J9U8 A0A0V1IPN2 A0A0J7KBA1 A0A0P6B8Y5 A0A0P6APU8
A0A1Y1K3Q7 A0A0V1M1K0 A0A0V0Z7R7 A0A1Y1LRT7 A0A0V1B5K0 A0A0V1B5A2 A0A0V1L945 A0A0J7K744 A0A0V0TK25 A0A0V0TK20 A0A0V1NHV8 J9LCG1 A0A139WFV5 A0A0V1HEX2 A0A2S2P896 A0A1Y1MYL8 A0A1Y1K2V0 A0A1Y1N6B6 X1WMG8 A0A1Y1MYL6 A0A1Y1NHZ0 A0A151ITN2 A0A0V0WE00 A0A0V0WA51 A0A0J7K9V7 A0A0V0W669 A0A1Y1KZM0 A0A0V1G2I5 X1XQX4 A0A2J7QWT7 A0A085N6E0 A0A0V1KG52 A0A0J7K763 X1XSG1 A0A0V1EYB7 A0A0J7N8C0 K7JBX1 K7J7F1 A0A0V1DTZ3 A0A0P5PQ39 K7JXE5 A0A1Y1NEM6 A0A1Y1L0K1 A0A0J7K2P0 A0A0J7K717 A0A0A9YZ05 A0A0A9WFQ0 K7JBT3 A0A085MSN1 A0A0J7K689 A0A1Y1LCN3 A0A0L0CMY6 K7JAW3 A0A0N5E2R3 A0A2A4JFW7 A0A0J7K651 X1WT66 A0A0J7KDE2 A0A1Y1KR09 A0A151J9U8 A0A0V1IPN2 A0A0J7KBA1 A0A0P6B8Y5 A0A0P6APU8
EMBL
AB378753
BAG68376.1
GDQN01003581
JAT87473.1
KK113102
KFM59342.1
+ More
KK112589 KFM58020.1 NEVH01023958 PNF17740.1 GEZM01078128 GEZM01078127 JAV63002.1 GEZM01099106 GEZM01099105 JAV53487.1 JYDO01000312 KRZ65659.1 JYDQ01000330 KRY08494.1 GEZM01048830 JAV76369.1 JYDH01000102 KRY32233.1 KRY32232.1 JYDW01000105 KRZ55879.1 LBMM01012672 KMQ86059.1 JYDJ01000235 KRX39370.1 KRX39371.1 JYDM01000206 KRZ83588.1 ABLF02020242 ABLF02020246 ABLF02042026 KQ971352 KYB26731.1 JYDP01000075 KRZ09245.1 GGMR01012789 MBY25408.1 GEZM01017328 JAV90763.1 GEZM01098859 JAV53706.1 GEZM01013586 GEZM01013585 JAV92450.1 ABLF02057951 ABLF02066512 GEZM01017329 JAV90762.1 GEZM01007166 GEZM01007165 JAV95327.1 KQ981003 KYN10396.1 JYDK01000164 KRX73801.1 JYDK01000219 KRX72067.1 LBMM01011114 KMQ87021.1 JYDK01000253 KRX71152.1 GEZM01073374 JAV65135.1 JYDT01000007 KRY92484.1 ABLF02042030 NEVH01009420 PNF33053.1 KL367545 KFD65036.1 JYDV01000001 KRZ46164.1 LBMM01012449 KMQ86217.1 ABLF02042028 JYDR01000003 KRY78530.1 LBMM01008433 KMQ88885.1 AAZX01004352 AAZX01012276 JYDR01000237 KRY65040.1 GDIQ01125974 JAL25752.1 AAZX01000127 GEZM01005616 JAV96038.1 JAV65136.1 LBMM01016284 KMQ84481.1 LBMM01012696 KMQ86054.1 GBHO01008809 JAG34795.1 GBHO01038256 JAG05348.1 AAZX01010628 AAZX01013403 KL367684 KFD60227.1 LBMM01012772 KMQ86003.1 GEZM01063091 JAV69316.1 JRES01000163 KNC33708.1 AAZX01023229 NWSH01001522 PCG70967.1 LBMM01012830 KMQ85968.1 ABLF02016448 LBMM01008998 KMQ88473.1 GEZM01078105 JAV63008.1 KQ979396 KYN21742.1 JYDV01000213 KRZ24707.1 LBMM01010133 KMQ87653.1 GDIP01019956 JAM83759.1 GDIP01039936 JAM63779.1
KK112589 KFM58020.1 NEVH01023958 PNF17740.1 GEZM01078128 GEZM01078127 JAV63002.1 GEZM01099106 GEZM01099105 JAV53487.1 JYDO01000312 KRZ65659.1 JYDQ01000330 KRY08494.1 GEZM01048830 JAV76369.1 JYDH01000102 KRY32233.1 KRY32232.1 JYDW01000105 KRZ55879.1 LBMM01012672 KMQ86059.1 JYDJ01000235 KRX39370.1 KRX39371.1 JYDM01000206 KRZ83588.1 ABLF02020242 ABLF02020246 ABLF02042026 KQ971352 KYB26731.1 JYDP01000075 KRZ09245.1 GGMR01012789 MBY25408.1 GEZM01017328 JAV90763.1 GEZM01098859 JAV53706.1 GEZM01013586 GEZM01013585 JAV92450.1 ABLF02057951 ABLF02066512 GEZM01017329 JAV90762.1 GEZM01007166 GEZM01007165 JAV95327.1 KQ981003 KYN10396.1 JYDK01000164 KRX73801.1 JYDK01000219 KRX72067.1 LBMM01011114 KMQ87021.1 JYDK01000253 KRX71152.1 GEZM01073374 JAV65135.1 JYDT01000007 KRY92484.1 ABLF02042030 NEVH01009420 PNF33053.1 KL367545 KFD65036.1 JYDV01000001 KRZ46164.1 LBMM01012449 KMQ86217.1 ABLF02042028 JYDR01000003 KRY78530.1 LBMM01008433 KMQ88885.1 AAZX01004352 AAZX01012276 JYDR01000237 KRY65040.1 GDIQ01125974 JAL25752.1 AAZX01000127 GEZM01005616 JAV96038.1 JAV65136.1 LBMM01016284 KMQ84481.1 LBMM01012696 KMQ86054.1 GBHO01008809 JAG34795.1 GBHO01038256 JAG05348.1 AAZX01010628 AAZX01013403 KL367684 KFD60227.1 LBMM01012772 KMQ86003.1 GEZM01063091 JAV69316.1 JRES01000163 KNC33708.1 AAZX01023229 NWSH01001522 PCG70967.1 LBMM01012830 KMQ85968.1 ABLF02016448 LBMM01008998 KMQ88473.1 GEZM01078105 JAV63008.1 KQ979396 KYN21742.1 JYDV01000213 KRZ24707.1 LBMM01010133 KMQ87653.1 GDIP01019956 JAM83759.1 GDIP01039936 JAM63779.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
B3Y003
A0A1E1WKD7
A0A087T2K3
A0A087SYT1
A0A2J7PN24
A0A1Y1KNM9
+ More
A0A1Y1K3Q7 A0A0V1M1K0 A0A0V0Z7R7 A0A1Y1LRT7 A0A0V1B5K0 A0A0V1B5A2 A0A0V1L945 A0A0J7K744 A0A0V0TK25 A0A0V0TK20 A0A0V1NHV8 J9LCG1 A0A139WFV5 A0A0V1HEX2 A0A2S2P896 A0A1Y1MYL8 A0A1Y1K2V0 A0A1Y1N6B6 X1WMG8 A0A1Y1MYL6 A0A1Y1NHZ0 A0A151ITN2 A0A0V0WE00 A0A0V0WA51 A0A0J7K9V7 A0A0V0W669 A0A1Y1KZM0 A0A0V1G2I5 X1XQX4 A0A2J7QWT7 A0A085N6E0 A0A0V1KG52 A0A0J7K763 X1XSG1 A0A0V1EYB7 A0A0J7N8C0 K7JBX1 K7J7F1 A0A0V1DTZ3 A0A0P5PQ39 K7JXE5 A0A1Y1NEM6 A0A1Y1L0K1 A0A0J7K2P0 A0A0J7K717 A0A0A9YZ05 A0A0A9WFQ0 K7JBT3 A0A085MSN1 A0A0J7K689 A0A1Y1LCN3 A0A0L0CMY6 K7JAW3 A0A0N5E2R3 A0A2A4JFW7 A0A0J7K651 X1WT66 A0A0J7KDE2 A0A1Y1KR09 A0A151J9U8 A0A0V1IPN2 A0A0J7KBA1 A0A0P6B8Y5 A0A0P6APU8
A0A1Y1K3Q7 A0A0V1M1K0 A0A0V0Z7R7 A0A1Y1LRT7 A0A0V1B5K0 A0A0V1B5A2 A0A0V1L945 A0A0J7K744 A0A0V0TK25 A0A0V0TK20 A0A0V1NHV8 J9LCG1 A0A139WFV5 A0A0V1HEX2 A0A2S2P896 A0A1Y1MYL8 A0A1Y1K2V0 A0A1Y1N6B6 X1WMG8 A0A1Y1MYL6 A0A1Y1NHZ0 A0A151ITN2 A0A0V0WE00 A0A0V0WA51 A0A0J7K9V7 A0A0V0W669 A0A1Y1KZM0 A0A0V1G2I5 X1XQX4 A0A2J7QWT7 A0A085N6E0 A0A0V1KG52 A0A0J7K763 X1XSG1 A0A0V1EYB7 A0A0J7N8C0 K7JBX1 K7J7F1 A0A0V1DTZ3 A0A0P5PQ39 K7JXE5 A0A1Y1NEM6 A0A1Y1L0K1 A0A0J7K2P0 A0A0J7K717 A0A0A9YZ05 A0A0A9WFQ0 K7JBT3 A0A085MSN1 A0A0J7K689 A0A1Y1LCN3 A0A0L0CMY6 K7JAW3 A0A0N5E2R3 A0A2A4JFW7 A0A0J7K651 X1WT66 A0A0J7KDE2 A0A1Y1KR09 A0A151J9U8 A0A0V1IPN2 A0A0J7KBA1 A0A0P6B8Y5 A0A0P6APU8
Ontologies
GO
PANTHER
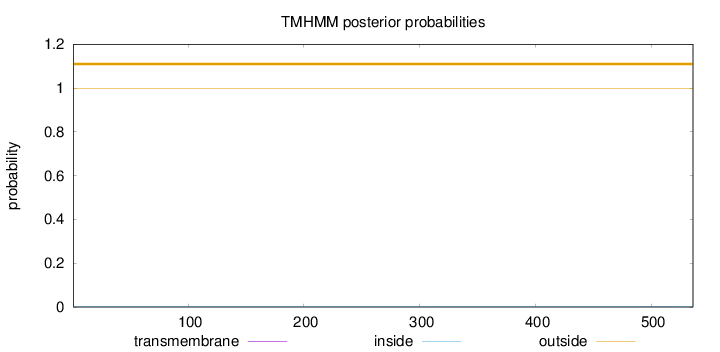
Topology
Length:
536
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00608
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.00100
outside
1 - 536
Population Genetic Test Statistics
Pi
8.403482
Theta
12.393585
Tajima's D
-0.952177
CLR
24.295354
CSRT
0.147492625368732
Interpretation
Uncertain
