Gene
KWMTBOMO04992 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008095
Annotation
short-chain_dehydrogenease/reductase-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.834
Sequence
CDS
ATGTTCGCTAATAAAGTTGTAATCATCACGGGAGCAAGTTCAGGAATAGGCGCTGAAACTGCATATGAATTCGCCAAATTGAATGCGAAATTGGTTTTAACTGGGAGAAAAAAGGAAAATCTTGATAATGTTGCATCCGATTGTGAAAGTCAATCTCCCAGTAAAATAAAACCGTTGGTAGTCGTGGCTGATATGAATAATGAGAGCGATGTCGAGAATATAATAAAAAAAACTGTTGATAACTATCACCAATTGGATGTGCTTGTAAATAATGCTGGTGTTTTGGAATCTGGTACCATAGAAACAACATCTTTGGCTCAATACGATAGAATAATGAACACTAATGTTCGCGGACCTTATTATTTGACGATGTTGGCAATTCCGCATTTGATTAAAACTAAAGGAAACATTGTCAACGTTTCCAGTGTTACTGGTTTGAGATCTTTCCCTAATGTCCTAGCGTATTGCATTTCTAAATCGGCAATAGATCAGTTTACAAGATGTGTGGCTTTAGAAGTAGCATTGAAAGGAGTTCGCGTGAATGCCGTCAACCCTGGGGTAATTACTACTGGTTTACATAAAAAAGGCGAAGGAAGTATGAACGAAGAACAATATGAAGCTTTTTTGGCAAAATGCGCAGAAACCCACGCTCTCGGCAGACCTGGGGATGCCAAAGAAGTATCTTCAGTCATCGTTTTCTTAGCTAGTGATGCTGCAAGCAACATTACTGGTGCCACTTTGCCTGTAGATGGAGGTCGTCATGCCATGTGTCCTAGATAA
Protein
MFANKVVIITGASSGIGAETAYEFAKLNAKLVLTGRKKENLDNVASDCESQSPSKIKPLVVVADMNNESDVENIIKKTVDNYHQLDVLVNNAGVLESGTIETTSLAQYDRIMNTNVRGPYYLTMLAIPHLIKTKGNIVNVSSVTGLRSFPNVLAYCISKSAIDQFTRCVALEVALKGVRVNAVNPGVITTGLHKKGEGSMNEEQYEAFLAKCAETHALGRPGDAKEVSSVIVFLASDAASNITGATLPVDGGRHAMCPR
Summary
Uniprot
H9JEZ8
Q2F651
A0A2H1WLI7
A0A0N1IEC4
A0A194QKD5
S4PS88
+ More
A0A2A4K3H7 A0A0L7KQD7 A0A212FE39 A0A2A4K3W4 H9JEZ9 A0A2H1WLM5 A0A194PR52 A0A194RFH9 S4PX81 S4P403 D6WBF1 H9IVP2 I4DNP9 Q2F652 A0A023ENP4 A0A182HAP4 J9HY30 A0A212F8M7 A0A2H1V849 A0A182LEW9 A0A2A4JMG8 A0A1W4XV70 A0A182XHV8 Q7PR84 A0A182HSU0 A0A182VPA1 A0A182TD91 B0X0M4 J3JT59 A0A2W1BPM7 U5EYA8 A0A1Q3FCK0 A0A182K3V2 A0A1S3IQP8 A0A182P965 A0A1I8PG95 E0VS61 A0A1L8EBS5 A0A1L8EDH6 A0A1L8EDA7 A0A182RS31 A0A1S3IQ98 A0A0M9A8J7 A0A2P8Y4R7 A0A1L8EDJ7 A0A2H8TRX4 A0A182TTS6 A0A1W7R596 A0A1J1IX96 A0A182V2E6 Q7QCA0 J9K4P0 R4FQ30 A0A1B6GU46 A0A2C9GR56 A0A026VV03 A0A1B6D562 A0A1B6JEH2 A0A1I8N405 A0A182M8T5 A0A0L0CQ82 A0A182SS86 A0A182LEW0 B4QWZ5 B4I447 A0A0L7KPW7 Q8IPP8 C4WUE4 A0A0J7KFU3 A0A1Y1LAV0 A0A1J1IYY0 W5J801 A0A023F8F0 A0A069DY41 Q8IGG7 A0A182Q2B3 A0A182IKA3 A0A084VXX9 A0A1L8EAQ3 B4PUV7 A0A2M4AJE9 A0A2M4AJQ0 A0A2M3Z8L5 Q170J0 A0A067R623 A0A2S2QKP0 T1DQK7 A0A0L7R3L7 A0A1S3D915 B4NKA4 A0A2M4BXH3 A0A2M4BY41 B3P2H5 A0A182FRI9
A0A2A4K3H7 A0A0L7KQD7 A0A212FE39 A0A2A4K3W4 H9JEZ9 A0A2H1WLM5 A0A194PR52 A0A194RFH9 S4PX81 S4P403 D6WBF1 H9IVP2 I4DNP9 Q2F652 A0A023ENP4 A0A182HAP4 J9HY30 A0A212F8M7 A0A2H1V849 A0A182LEW9 A0A2A4JMG8 A0A1W4XV70 A0A182XHV8 Q7PR84 A0A182HSU0 A0A182VPA1 A0A182TD91 B0X0M4 J3JT59 A0A2W1BPM7 U5EYA8 A0A1Q3FCK0 A0A182K3V2 A0A1S3IQP8 A0A182P965 A0A1I8PG95 E0VS61 A0A1L8EBS5 A0A1L8EDH6 A0A1L8EDA7 A0A182RS31 A0A1S3IQ98 A0A0M9A8J7 A0A2P8Y4R7 A0A1L8EDJ7 A0A2H8TRX4 A0A182TTS6 A0A1W7R596 A0A1J1IX96 A0A182V2E6 Q7QCA0 J9K4P0 R4FQ30 A0A1B6GU46 A0A2C9GR56 A0A026VV03 A0A1B6D562 A0A1B6JEH2 A0A1I8N405 A0A182M8T5 A0A0L0CQ82 A0A182SS86 A0A182LEW0 B4QWZ5 B4I447 A0A0L7KPW7 Q8IPP8 C4WUE4 A0A0J7KFU3 A0A1Y1LAV0 A0A1J1IYY0 W5J801 A0A023F8F0 A0A069DY41 Q8IGG7 A0A182Q2B3 A0A182IKA3 A0A084VXX9 A0A1L8EAQ3 B4PUV7 A0A2M4AJE9 A0A2M4AJQ0 A0A2M3Z8L5 Q170J0 A0A067R623 A0A2S2QKP0 T1DQK7 A0A0L7R3L7 A0A1S3D915 B4NKA4 A0A2M4BXH3 A0A2M4BY41 B3P2H5 A0A182FRI9
Pubmed
19121390
26354079
23622113
26227816
22118469
18362917
+ More
19820115 22651552 24945155 26483478 17510324 20966253 12364791 14747013 17210077 22516182 23537049 28756777 20566863 29403074 24508170 30249741 25315136 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 28004739 20920257 23761445 25474469 26334808 24438588 17550304 17519023 24845553
19820115 22651552 24945155 26483478 17510324 20966253 12364791 14747013 17210077 22516182 23537049 28756777 20566863 29403074 24508170 30249741 25315136 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 28004739 20920257 23761445 25474469 26334808 24438588 17550304 17519023 24845553
EMBL
BABH01033917
DQ311221
ABD36166.1
ODYU01009467
SOQ53929.1
KQ458665
+ More
KPJ05623.1 KQ461198 KPJ06033.1 GAIX01000150 JAA92410.1 NWSH01000203 PCG78454.1 JTDY01007021 KOB65518.1 AGBW02008979 OWR51994.1 PCG78453.1 SOQ53928.1 KQ459595 KPI95931.1 KQ460313 KPJ16055.1 GAIX01005498 JAA87062.1 GAIX01006214 JAA86346.1 KQ971312 EEZ97890.1 BABH01039662 AK403072 BAM19539.1 DQ311220 ABD36165.1 GAPW01003549 JAC10049.1 JXUM01081382 JXUM01123063 KQ566675 KQ563243 KXJ69914.1 KXJ74217.1 CH477221 EJY57385.1 AGBW02009702 OWR50087.1 ODYU01001173 SOQ37020.1 NWSH01000983 PCG73247.1 AAAB01008859 EAA07681.6 APCN01000350 DS232242 EDS38266.1 BT126415 KB632402 AEE61379.1 ERL94969.1 KZ149964 PZC76241.1 GANO01000477 JAB59394.1 GFDL01009740 JAV25305.1 DS235745 EEB16217.1 GFDG01002611 JAV16188.1 GFDG01002077 JAV16722.1 GFDG01002078 JAV16721.1 KQ435726 KOX78123.1 PYGN01000933 PSN39247.1 GFDG01002075 JAV16724.1 GFXV01005148 MBW16953.1 GEHC01001354 JAV46291.1 CVRI01000063 CRL04899.1 EAA07469.4 EGK96869.1 ABLF02005808 ACPB03001903 GAHY01000619 JAA76891.1 GECZ01003828 JAS65941.1 KK107871 QOIP01000007 EZA47366.1 RLU20203.1 GEDC01016505 JAS20793.1 GECU01010179 JAS97527.1 AXCM01010093 JRES01000061 KNC34518.1 CM000364 EDX11757.1 CH480821 EDW54990.1 JTDY01007683 KOB65019.1 AE014297 AAN13253.1 AK341030 BAH71514.1 LBMM01007925 KMQ89303.1 GEZM01064587 JAV68706.1 CRL04898.1 ADMH02002105 ETN58985.1 GBBI01001127 GEMB01003820 JAC17585.1 JAR99434.1 GBGD01002600 JAC86289.1 BT001792 AAN71547.1 AXCN02000007 ATLV01018212 KE525224 KFB42823.1 GFDG01002982 JAV15817.1 CM000160 EDW97764.1 GGFK01007592 MBW40913.1 GGFK01007692 MBW41013.1 GGFM01004116 MBW24867.1 EF173379 CH477473 ABM68625.1 EAT40351.1 KK852678 KDR18660.1 GGMS01008897 MBY78100.1 GAMD01001105 JAB00486.1 KQ414663 KOC65477.1 CH964272 EDW84034.1 GGFJ01008571 MBW57712.1 GGFJ01008517 MBW57658.1 CH954181 EDV47925.1
KPJ05623.1 KQ461198 KPJ06033.1 GAIX01000150 JAA92410.1 NWSH01000203 PCG78454.1 JTDY01007021 KOB65518.1 AGBW02008979 OWR51994.1 PCG78453.1 SOQ53928.1 KQ459595 KPI95931.1 KQ460313 KPJ16055.1 GAIX01005498 JAA87062.1 GAIX01006214 JAA86346.1 KQ971312 EEZ97890.1 BABH01039662 AK403072 BAM19539.1 DQ311220 ABD36165.1 GAPW01003549 JAC10049.1 JXUM01081382 JXUM01123063 KQ566675 KQ563243 KXJ69914.1 KXJ74217.1 CH477221 EJY57385.1 AGBW02009702 OWR50087.1 ODYU01001173 SOQ37020.1 NWSH01000983 PCG73247.1 AAAB01008859 EAA07681.6 APCN01000350 DS232242 EDS38266.1 BT126415 KB632402 AEE61379.1 ERL94969.1 KZ149964 PZC76241.1 GANO01000477 JAB59394.1 GFDL01009740 JAV25305.1 DS235745 EEB16217.1 GFDG01002611 JAV16188.1 GFDG01002077 JAV16722.1 GFDG01002078 JAV16721.1 KQ435726 KOX78123.1 PYGN01000933 PSN39247.1 GFDG01002075 JAV16724.1 GFXV01005148 MBW16953.1 GEHC01001354 JAV46291.1 CVRI01000063 CRL04899.1 EAA07469.4 EGK96869.1 ABLF02005808 ACPB03001903 GAHY01000619 JAA76891.1 GECZ01003828 JAS65941.1 KK107871 QOIP01000007 EZA47366.1 RLU20203.1 GEDC01016505 JAS20793.1 GECU01010179 JAS97527.1 AXCM01010093 JRES01000061 KNC34518.1 CM000364 EDX11757.1 CH480821 EDW54990.1 JTDY01007683 KOB65019.1 AE014297 AAN13253.1 AK341030 BAH71514.1 LBMM01007925 KMQ89303.1 GEZM01064587 JAV68706.1 CRL04898.1 ADMH02002105 ETN58985.1 GBBI01001127 GEMB01003820 JAC17585.1 JAR99434.1 GBGD01002600 JAC86289.1 BT001792 AAN71547.1 AXCN02000007 ATLV01018212 KE525224 KFB42823.1 GFDG01002982 JAV15817.1 CM000160 EDW97764.1 GGFK01007592 MBW40913.1 GGFK01007692 MBW41013.1 GGFM01004116 MBW24867.1 EF173379 CH477473 ABM68625.1 EAT40351.1 KK852678 KDR18660.1 GGMS01008897 MBY78100.1 GAMD01001105 JAB00486.1 KQ414663 KOC65477.1 CH964272 EDW84034.1 GGFJ01008571 MBW57712.1 GGFJ01008517 MBW57658.1 CH954181 EDV47925.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000007151
+ More
UP000007266 UP000069940 UP000249989 UP000008820 UP000075882 UP000192223 UP000076407 UP000007062 UP000075840 UP000075903 UP000075902 UP000002320 UP000030742 UP000075881 UP000085678 UP000075885 UP000095300 UP000009046 UP000075900 UP000053105 UP000245037 UP000183832 UP000007819 UP000015103 UP000053097 UP000279307 UP000095301 UP000075883 UP000037069 UP000075901 UP000000304 UP000001292 UP000000803 UP000036403 UP000000673 UP000075886 UP000075880 UP000030765 UP000002282 UP000027135 UP000053825 UP000079169 UP000007798 UP000008711 UP000069272
UP000007266 UP000069940 UP000249989 UP000008820 UP000075882 UP000192223 UP000076407 UP000007062 UP000075840 UP000075903 UP000075902 UP000002320 UP000030742 UP000075881 UP000085678 UP000075885 UP000095300 UP000009046 UP000075900 UP000053105 UP000245037 UP000183832 UP000007819 UP000015103 UP000053097 UP000279307 UP000095301 UP000075883 UP000037069 UP000075901 UP000000304 UP000001292 UP000000803 UP000036403 UP000000673 UP000075886 UP000075880 UP000030765 UP000002282 UP000027135 UP000053825 UP000079169 UP000007798 UP000008711 UP000069272
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JEZ8
Q2F651
A0A2H1WLI7
A0A0N1IEC4
A0A194QKD5
S4PS88
+ More
A0A2A4K3H7 A0A0L7KQD7 A0A212FE39 A0A2A4K3W4 H9JEZ9 A0A2H1WLM5 A0A194PR52 A0A194RFH9 S4PX81 S4P403 D6WBF1 H9IVP2 I4DNP9 Q2F652 A0A023ENP4 A0A182HAP4 J9HY30 A0A212F8M7 A0A2H1V849 A0A182LEW9 A0A2A4JMG8 A0A1W4XV70 A0A182XHV8 Q7PR84 A0A182HSU0 A0A182VPA1 A0A182TD91 B0X0M4 J3JT59 A0A2W1BPM7 U5EYA8 A0A1Q3FCK0 A0A182K3V2 A0A1S3IQP8 A0A182P965 A0A1I8PG95 E0VS61 A0A1L8EBS5 A0A1L8EDH6 A0A1L8EDA7 A0A182RS31 A0A1S3IQ98 A0A0M9A8J7 A0A2P8Y4R7 A0A1L8EDJ7 A0A2H8TRX4 A0A182TTS6 A0A1W7R596 A0A1J1IX96 A0A182V2E6 Q7QCA0 J9K4P0 R4FQ30 A0A1B6GU46 A0A2C9GR56 A0A026VV03 A0A1B6D562 A0A1B6JEH2 A0A1I8N405 A0A182M8T5 A0A0L0CQ82 A0A182SS86 A0A182LEW0 B4QWZ5 B4I447 A0A0L7KPW7 Q8IPP8 C4WUE4 A0A0J7KFU3 A0A1Y1LAV0 A0A1J1IYY0 W5J801 A0A023F8F0 A0A069DY41 Q8IGG7 A0A182Q2B3 A0A182IKA3 A0A084VXX9 A0A1L8EAQ3 B4PUV7 A0A2M4AJE9 A0A2M4AJQ0 A0A2M3Z8L5 Q170J0 A0A067R623 A0A2S2QKP0 T1DQK7 A0A0L7R3L7 A0A1S3D915 B4NKA4 A0A2M4BXH3 A0A2M4BY41 B3P2H5 A0A182FRI9
A0A2A4K3H7 A0A0L7KQD7 A0A212FE39 A0A2A4K3W4 H9JEZ9 A0A2H1WLM5 A0A194PR52 A0A194RFH9 S4PX81 S4P403 D6WBF1 H9IVP2 I4DNP9 Q2F652 A0A023ENP4 A0A182HAP4 J9HY30 A0A212F8M7 A0A2H1V849 A0A182LEW9 A0A2A4JMG8 A0A1W4XV70 A0A182XHV8 Q7PR84 A0A182HSU0 A0A182VPA1 A0A182TD91 B0X0M4 J3JT59 A0A2W1BPM7 U5EYA8 A0A1Q3FCK0 A0A182K3V2 A0A1S3IQP8 A0A182P965 A0A1I8PG95 E0VS61 A0A1L8EBS5 A0A1L8EDH6 A0A1L8EDA7 A0A182RS31 A0A1S3IQ98 A0A0M9A8J7 A0A2P8Y4R7 A0A1L8EDJ7 A0A2H8TRX4 A0A182TTS6 A0A1W7R596 A0A1J1IX96 A0A182V2E6 Q7QCA0 J9K4P0 R4FQ30 A0A1B6GU46 A0A2C9GR56 A0A026VV03 A0A1B6D562 A0A1B6JEH2 A0A1I8N405 A0A182M8T5 A0A0L0CQ82 A0A182SS86 A0A182LEW0 B4QWZ5 B4I447 A0A0L7KPW7 Q8IPP8 C4WUE4 A0A0J7KFU3 A0A1Y1LAV0 A0A1J1IYY0 W5J801 A0A023F8F0 A0A069DY41 Q8IGG7 A0A182Q2B3 A0A182IKA3 A0A084VXX9 A0A1L8EAQ3 B4PUV7 A0A2M4AJE9 A0A2M4AJQ0 A0A2M3Z8L5 Q170J0 A0A067R623 A0A2S2QKP0 T1DQK7 A0A0L7R3L7 A0A1S3D915 B4NKA4 A0A2M4BXH3 A0A2M4BY41 B3P2H5 A0A182FRI9
PDB
1SPX
E-value=1.2394e-37,
Score=390
Ontologies
GO
Topology
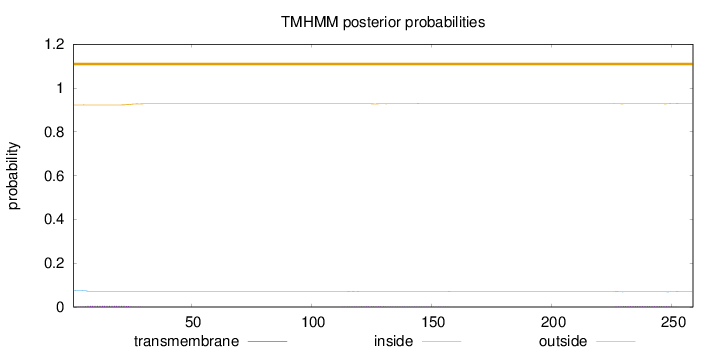
Length:
259
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21494
Exp number, first 60 AAs:
0.1125
Total prob of N-in:
0.07694
outside
1 - 259
Population Genetic Test Statistics
Pi
27.002811
Theta
24.125414
Tajima's D
0.307129
CLR
296.109837
CSRT
0.467776611169442
Interpretation
Uncertain
