Gene
KWMTBOMO04978
Annotation
pyruvate_kinase_[Danaus_plexippus]
Full name
Pyruvate kinase
Location in the cell
Mitochondrial Reliability : 1.498
Sequence
CDS
ATGATAACTCGACAGGAATTAGGTTCTGATATAACACCGAAGAAGCTAGTCATTGCTCAGAAGAATATGATTGCACGTGCGAATAAGGCTAACGTTCCGGTCTGTGTCAACGCACATCTCCTAAGCAGCATGAGATACAAAAAATTACCACTAAGAGCGGAACTTTTGGATGTGGCGAACTGCATTCTGGATGGTGCCGACACTCTTGAATTATCTGCTGAAACAGCGGTCGGTCTGTACCCAGTCGACACTGTTGCCTGTTTAGCCAGTGCCTGTAAAGAAGCTGAAGCCTGCATCTGGTCTAAGCAGATTTTTCACGATTTTGTCGATAAGACGCCGATCCCGTGCGATCAAGCAACCGGTGTTGGATTAGCCGCGGTACTGGCAGCGCAGCGGTCATTAGCGGCCGCCATAATTGTCGTCACCACAACCGGGAAATCGGCGCAAATTGTATCTAAATACGGACCGAGATGTCCCATTATAGGTGTAACTAGATACGCTGCTATAGCTAGACAATTGCACTTGTGGAGAGGAATCATACCGCTTATTTTTGAGGGTACTTAA
Protein
MITRQELGSDITPKKLVIAQKNMIARANKANVPVCVNAHLLSSMRYKKLPLRAELLDVANCILDGADTLELSAETAVGLYPVDTVACLASACKEAEACIWSKQIFHDFVDKTPIPCDQATGVGLAAVLAAQRSLAAAIIVVTTTGKSAQIVSKYGPRCPIIGVTRYAAIARQLHLWRGIIPLIFEGT
Summary
Catalytic Activity
ATP + pyruvate = ADP + H(+) + phosphoenolpyruvate
Similarity
Belongs to the pyruvate kinase family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Uniprot
A0A212FH64
A0A194PDF2
A0A2A4JE60
A0A194QKG4
A0A2H1V847
A0A0L7L0B6
+ More
A0A1B6MST3 A0A182SYI1 A0A1Y9GJY2 Q7PPE7 A0A1Y9GJK0 A0A084W7S0 A0A182UE55 D6WXE6 A0A139WDM5 A0A182NN15 A0A182RED7 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A1B6KVA1 A0A182VFI7 A0A182LNM2 A0A182QGD3 A0A182KAF7 A0A182FAJ1 A0A2M4CU89 A0A182MNQ6 A0A182JJQ1 A0A2M3ZYS8 A0A182VPN6 A0A2M3ZYS6 T1DN39 W5JLV6 A0A182PWA9 A0A182YG83 A0A2M3YXR0 A0A2M3ZFB9 Q16LP4 A0A2M3YXN9 Q16LP5 A0A0J7K3Z0 Q16F38 A0A0J7NMP8 E9J2S7 A0A1B6CT72 A0A1B6DMR8 A0A023EVC5 E2AJ75 A0A240SY09 A0A026WCK1 A0A158NID4 A0A154PSS0 A0A151HZ28 A0A151XDE9 A0A151J4F0 A0A151JZ74 H9IZ23 A0A3L8DTT9 A0A1Q3FL43 A0A195C2R8 B0WL21 A0A1Q3FKT7 A0A1Q3FKV4 A0A212FLH1 A0A194QBK5 A0A1Q3FL39 W8BPB9 A0A1B6GSY1 A0A1L8E1V8 A0A182H1Q2 A0A1B6GQK1 W8BUX5 A0A0K8TLX8 A0A2R7VZN0 A0A1L8E288 A0A1X9QDT2 A0A1W5YLK4 E2BFQ8 A0A1B6F736 A0A2Z5REH9 A0A226EGV1 R4WDM9 A0A0K8SRD7 A0A0K8SR30 A0A0A9XHK7 A0A0A9YKR8 A0A2Z5REH8 F4WXV2 A0A2H1V6Z8 A0A067R9U1 A0A0A9YVU7 A0A0L7QJZ6 U5ESC2 A0A0N1ITM9 I4DJQ4 A0A1I8NY84 A0A1D2NHX8 A0A1I8NY83 A0A170YTJ4
A0A1B6MST3 A0A182SYI1 A0A1Y9GJY2 Q7PPE7 A0A1Y9GJK0 A0A084W7S0 A0A182UE55 D6WXE6 A0A139WDM5 A0A182NN15 A0A182RED7 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A1B6KVA1 A0A182VFI7 A0A182LNM2 A0A182QGD3 A0A182KAF7 A0A182FAJ1 A0A2M4CU89 A0A182MNQ6 A0A182JJQ1 A0A2M3ZYS8 A0A182VPN6 A0A2M3ZYS6 T1DN39 W5JLV6 A0A182PWA9 A0A182YG83 A0A2M3YXR0 A0A2M3ZFB9 Q16LP4 A0A2M3YXN9 Q16LP5 A0A0J7K3Z0 Q16F38 A0A0J7NMP8 E9J2S7 A0A1B6CT72 A0A1B6DMR8 A0A023EVC5 E2AJ75 A0A240SY09 A0A026WCK1 A0A158NID4 A0A154PSS0 A0A151HZ28 A0A151XDE9 A0A151J4F0 A0A151JZ74 H9IZ23 A0A3L8DTT9 A0A1Q3FL43 A0A195C2R8 B0WL21 A0A1Q3FKT7 A0A1Q3FKV4 A0A212FLH1 A0A194QBK5 A0A1Q3FL39 W8BPB9 A0A1B6GSY1 A0A1L8E1V8 A0A182H1Q2 A0A1B6GQK1 W8BUX5 A0A0K8TLX8 A0A2R7VZN0 A0A1L8E288 A0A1X9QDT2 A0A1W5YLK4 E2BFQ8 A0A1B6F736 A0A2Z5REH9 A0A226EGV1 R4WDM9 A0A0K8SRD7 A0A0K8SR30 A0A0A9XHK7 A0A0A9YKR8 A0A2Z5REH8 F4WXV2 A0A2H1V6Z8 A0A067R9U1 A0A0A9YVU7 A0A0L7QJZ6 U5ESC2 A0A0N1ITM9 I4DJQ4 A0A1I8NY84 A0A1D2NHX8 A0A1I8NY83 A0A170YTJ4
EC Number
2.7.1.40
Pubmed
EMBL
AGBW02008537
OWR53070.1
KQ459606
KPI91277.1
NWSH01001903
PCG69730.1
+ More
KQ461198 KPJ06022.1 ODYU01000922 SOQ36414.1 JTDY01003837 KOB68932.1 GEBQ01001009 JAT38968.1 APCN01005967 AAAB01008952 EAA10555.6 ATLV01021293 KE525316 KFB46264.1 KQ971361 EFA08832.2 KYB25941.1 GEBQ01024610 JAT15367.1 AXCN02001590 GGFL01004734 MBW68912.1 AXCM01000245 GGFK01000396 MBW33717.1 GGFK01000395 MBW33716.1 GAMD01003210 JAA98380.1 ADMH02000764 GGFL01004376 ETN65121.1 MBW68554.1 GGFM01000290 MBW21041.1 GGFM01006434 MBW27185.1 CH477898 EAT35243.1 GGFM01000291 MBW21042.1 EAT35242.1 LBMM01014568 KMQ85133.1 CH478516 EAT32846.1 LBMM01003236 KMQ93775.1 GL768000 EFZ12877.1 GEDC01020559 JAS16739.1 GEDC01010333 JAS26965.1 GAPW01000661 JAC12937.1 GL439967 EFN66515.1 AJWK01004665 AJWK01004666 AJWK01004667 KK107293 EZA53406.1 ADTU01016559 ADTU01016560 ADTU01016561 ADTU01016562 ADTU01016563 ADTU01016564 KQ435161 KZC14923.1 KQ976702 KYM77205.1 KQ982294 KYQ58406.1 KQ980157 KYN17435.1 KQ981432 KYN41677.1 BABH01010950 BABH01010951 BABH01010952 QOIP01000005 RLU23178.1 GFDL01006821 JAV28224.1 KQ978379 KYM94483.1 DS231979 EDS30184.1 GFDL01006824 JAV28221.1 GFDL01006857 JAV28188.1 AGBW02007775 OWR54577.1 KQ459220 KPJ02928.1 GFDL01006859 JAV28186.1 GAMC01005788 JAC00768.1 GECZ01004238 JAS65531.1 GFDF01001448 JAV12636.1 JXUM01023189 JXUM01023190 KQ560654 KXJ81422.1 GECZ01005099 JAS64670.1 GAMC01005787 JAC00769.1 GDAI01002239 JAI15364.1 KK854197 PTY12947.1 GFDF01001335 JAV12749.1 KY412773 ARQ20739.1 KX147661 ARI45065.1 GL448039 EFN85446.1 GECZ01024076 JAS45693.1 FX983728 BAX07227.1 LNIX01000004 OXA56448.1 AK417674 BAN20889.1 GBRD01012428 GBRD01012427 GBRD01012426 GBRD01012425 GBRD01012424 GBRD01012422 GBRD01010042 GBRD01010040 GBRD01010039 GBRD01010038 GBRD01006700 GBRD01006699 JAG55786.1 GBRD01010044 GBRD01010041 GBRD01002395 JAG55783.1 GBHO01024498 GBHO01023780 GBHO01010803 GBHO01010230 GBHO01006495 GBHO01006493 JAG19106.1 JAG19824.1 JAG32801.1 JAG33374.1 JAG37109.1 JAG37111.1 GBHO01010825 GBHO01010824 GBHO01010805 GBHO01010307 GBHO01010289 GBHO01009918 GDHC01007241 JAG32779.1 JAG32780.1 JAG32799.1 JAG33297.1 JAG33315.1 JAG33686.1 JAQ11388.1 FX983727 BAX07226.1 GL888432 EGI60975.1 ODYU01000813 SOQ36162.1 KK852651 KDR19430.1 GBHO01023781 GBHO01023776 GBHO01023775 GBHO01010806 GBHO01009950 GBHO01006502 GBHO01006497 JAG19823.1 JAG19828.1 JAG19829.1 JAG32798.1 JAG33654.1 JAG37102.1 JAG37107.1 KQ415008 KOC58948.1 GANO01002423 JAB57448.1 KQ435775 KOX74974.1 AK401522 BAM18144.1 LJIJ01000034 ODN04873.1 GEMB01002976 JAS00227.1
KQ461198 KPJ06022.1 ODYU01000922 SOQ36414.1 JTDY01003837 KOB68932.1 GEBQ01001009 JAT38968.1 APCN01005967 AAAB01008952 EAA10555.6 ATLV01021293 KE525316 KFB46264.1 KQ971361 EFA08832.2 KYB25941.1 GEBQ01024610 JAT15367.1 AXCN02001590 GGFL01004734 MBW68912.1 AXCM01000245 GGFK01000396 MBW33717.1 GGFK01000395 MBW33716.1 GAMD01003210 JAA98380.1 ADMH02000764 GGFL01004376 ETN65121.1 MBW68554.1 GGFM01000290 MBW21041.1 GGFM01006434 MBW27185.1 CH477898 EAT35243.1 GGFM01000291 MBW21042.1 EAT35242.1 LBMM01014568 KMQ85133.1 CH478516 EAT32846.1 LBMM01003236 KMQ93775.1 GL768000 EFZ12877.1 GEDC01020559 JAS16739.1 GEDC01010333 JAS26965.1 GAPW01000661 JAC12937.1 GL439967 EFN66515.1 AJWK01004665 AJWK01004666 AJWK01004667 KK107293 EZA53406.1 ADTU01016559 ADTU01016560 ADTU01016561 ADTU01016562 ADTU01016563 ADTU01016564 KQ435161 KZC14923.1 KQ976702 KYM77205.1 KQ982294 KYQ58406.1 KQ980157 KYN17435.1 KQ981432 KYN41677.1 BABH01010950 BABH01010951 BABH01010952 QOIP01000005 RLU23178.1 GFDL01006821 JAV28224.1 KQ978379 KYM94483.1 DS231979 EDS30184.1 GFDL01006824 JAV28221.1 GFDL01006857 JAV28188.1 AGBW02007775 OWR54577.1 KQ459220 KPJ02928.1 GFDL01006859 JAV28186.1 GAMC01005788 JAC00768.1 GECZ01004238 JAS65531.1 GFDF01001448 JAV12636.1 JXUM01023189 JXUM01023190 KQ560654 KXJ81422.1 GECZ01005099 JAS64670.1 GAMC01005787 JAC00769.1 GDAI01002239 JAI15364.1 KK854197 PTY12947.1 GFDF01001335 JAV12749.1 KY412773 ARQ20739.1 KX147661 ARI45065.1 GL448039 EFN85446.1 GECZ01024076 JAS45693.1 FX983728 BAX07227.1 LNIX01000004 OXA56448.1 AK417674 BAN20889.1 GBRD01012428 GBRD01012427 GBRD01012426 GBRD01012425 GBRD01012424 GBRD01012422 GBRD01010042 GBRD01010040 GBRD01010039 GBRD01010038 GBRD01006700 GBRD01006699 JAG55786.1 GBRD01010044 GBRD01010041 GBRD01002395 JAG55783.1 GBHO01024498 GBHO01023780 GBHO01010803 GBHO01010230 GBHO01006495 GBHO01006493 JAG19106.1 JAG19824.1 JAG32801.1 JAG33374.1 JAG37109.1 JAG37111.1 GBHO01010825 GBHO01010824 GBHO01010805 GBHO01010307 GBHO01010289 GBHO01009918 GDHC01007241 JAG32779.1 JAG32780.1 JAG32799.1 JAG33297.1 JAG33315.1 JAG33686.1 JAQ11388.1 FX983727 BAX07226.1 GL888432 EGI60975.1 ODYU01000813 SOQ36162.1 KK852651 KDR19430.1 GBHO01023781 GBHO01023776 GBHO01023775 GBHO01010806 GBHO01009950 GBHO01006502 GBHO01006497 JAG19823.1 JAG19828.1 JAG19829.1 JAG32798.1 JAG33654.1 JAG37102.1 JAG37107.1 KQ415008 KOC58948.1 GANO01002423 JAB57448.1 KQ435775 KOX74974.1 AK401522 BAM18144.1 LJIJ01000034 ODN04873.1 GEMB01002976 JAS00227.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000053240
UP000037510
UP000075901
+ More
UP000075840 UP000007062 UP000030765 UP000075902 UP000007266 UP000075884 UP000075900 UP000076407 UP000075903 UP000075882 UP000075886 UP000075881 UP000069272 UP000075883 UP000075880 UP000075920 UP000000673 UP000075885 UP000076408 UP000008820 UP000036403 UP000000311 UP000092461 UP000053097 UP000005205 UP000076502 UP000078540 UP000075809 UP000078492 UP000078541 UP000005204 UP000279307 UP000078542 UP000002320 UP000069940 UP000249989 UP000008237 UP000198287 UP000007755 UP000027135 UP000053825 UP000053105 UP000095300 UP000094527
UP000075840 UP000007062 UP000030765 UP000075902 UP000007266 UP000075884 UP000075900 UP000076407 UP000075903 UP000075882 UP000075886 UP000075881 UP000069272 UP000075883 UP000075880 UP000075920 UP000000673 UP000075885 UP000076408 UP000008820 UP000036403 UP000000311 UP000092461 UP000053097 UP000005205 UP000076502 UP000078540 UP000075809 UP000078492 UP000078541 UP000005204 UP000279307 UP000078542 UP000002320 UP000069940 UP000249989 UP000008237 UP000198287 UP000007755 UP000027135 UP000053825 UP000053105 UP000095300 UP000094527
PRIDE
Interpro
IPR011037
Pyrv_Knase-like_insert_dom_sf
+ More
IPR015806 Pyrv_Knase_insert_dom_sf
IPR036918 Pyrv_Knase_C_sf
IPR040442 Pyrv_Kinase-like_dom_sf
IPR015813 Pyrv/PenolPyrv_Kinase-like_dom
IPR001697 Pyr_Knase
IPR015795 Pyrv_Knase_C
IPR015793 Pyrv_Knase_brl
IPR018209 Pyrv_Knase_AS
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR001737 KsgA/Erm
IPR029063 SAM-dependent_MTases
IPR015806 Pyrv_Knase_insert_dom_sf
IPR036918 Pyrv_Knase_C_sf
IPR040442 Pyrv_Kinase-like_dom_sf
IPR015813 Pyrv/PenolPyrv_Kinase-like_dom
IPR001697 Pyr_Knase
IPR015795 Pyrv_Knase_C
IPR015793 Pyrv_Knase_brl
IPR018209 Pyrv_Knase_AS
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR001737 KsgA/Erm
IPR029063 SAM-dependent_MTases
Gene 3D
ProteinModelPortal
A0A212FH64
A0A194PDF2
A0A2A4JE60
A0A194QKG4
A0A2H1V847
A0A0L7L0B6
+ More
A0A1B6MST3 A0A182SYI1 A0A1Y9GJY2 Q7PPE7 A0A1Y9GJK0 A0A084W7S0 A0A182UE55 D6WXE6 A0A139WDM5 A0A182NN15 A0A182RED7 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A1B6KVA1 A0A182VFI7 A0A182LNM2 A0A182QGD3 A0A182KAF7 A0A182FAJ1 A0A2M4CU89 A0A182MNQ6 A0A182JJQ1 A0A2M3ZYS8 A0A182VPN6 A0A2M3ZYS6 T1DN39 W5JLV6 A0A182PWA9 A0A182YG83 A0A2M3YXR0 A0A2M3ZFB9 Q16LP4 A0A2M3YXN9 Q16LP5 A0A0J7K3Z0 Q16F38 A0A0J7NMP8 E9J2S7 A0A1B6CT72 A0A1B6DMR8 A0A023EVC5 E2AJ75 A0A240SY09 A0A026WCK1 A0A158NID4 A0A154PSS0 A0A151HZ28 A0A151XDE9 A0A151J4F0 A0A151JZ74 H9IZ23 A0A3L8DTT9 A0A1Q3FL43 A0A195C2R8 B0WL21 A0A1Q3FKT7 A0A1Q3FKV4 A0A212FLH1 A0A194QBK5 A0A1Q3FL39 W8BPB9 A0A1B6GSY1 A0A1L8E1V8 A0A182H1Q2 A0A1B6GQK1 W8BUX5 A0A0K8TLX8 A0A2R7VZN0 A0A1L8E288 A0A1X9QDT2 A0A1W5YLK4 E2BFQ8 A0A1B6F736 A0A2Z5REH9 A0A226EGV1 R4WDM9 A0A0K8SRD7 A0A0K8SR30 A0A0A9XHK7 A0A0A9YKR8 A0A2Z5REH8 F4WXV2 A0A2H1V6Z8 A0A067R9U1 A0A0A9YVU7 A0A0L7QJZ6 U5ESC2 A0A0N1ITM9 I4DJQ4 A0A1I8NY84 A0A1D2NHX8 A0A1I8NY83 A0A170YTJ4
A0A1B6MST3 A0A182SYI1 A0A1Y9GJY2 Q7PPE7 A0A1Y9GJK0 A0A084W7S0 A0A182UE55 D6WXE6 A0A139WDM5 A0A182NN15 A0A182RED7 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A1B6KVA1 A0A182VFI7 A0A182LNM2 A0A182QGD3 A0A182KAF7 A0A182FAJ1 A0A2M4CU89 A0A182MNQ6 A0A182JJQ1 A0A2M3ZYS8 A0A182VPN6 A0A2M3ZYS6 T1DN39 W5JLV6 A0A182PWA9 A0A182YG83 A0A2M3YXR0 A0A2M3ZFB9 Q16LP4 A0A2M3YXN9 Q16LP5 A0A0J7K3Z0 Q16F38 A0A0J7NMP8 E9J2S7 A0A1B6CT72 A0A1B6DMR8 A0A023EVC5 E2AJ75 A0A240SY09 A0A026WCK1 A0A158NID4 A0A154PSS0 A0A151HZ28 A0A151XDE9 A0A151J4F0 A0A151JZ74 H9IZ23 A0A3L8DTT9 A0A1Q3FL43 A0A195C2R8 B0WL21 A0A1Q3FKT7 A0A1Q3FKV4 A0A212FLH1 A0A194QBK5 A0A1Q3FL39 W8BPB9 A0A1B6GSY1 A0A1L8E1V8 A0A182H1Q2 A0A1B6GQK1 W8BUX5 A0A0K8TLX8 A0A2R7VZN0 A0A1L8E288 A0A1X9QDT2 A0A1W5YLK4 E2BFQ8 A0A1B6F736 A0A2Z5REH9 A0A226EGV1 R4WDM9 A0A0K8SRD7 A0A0K8SR30 A0A0A9XHK7 A0A0A9YKR8 A0A2Z5REH8 F4WXV2 A0A2H1V6Z8 A0A067R9U1 A0A0A9YVU7 A0A0L7QJZ6 U5ESC2 A0A0N1ITM9 I4DJQ4 A0A1I8NY84 A0A1D2NHX8 A0A1I8NY83 A0A170YTJ4
PDB
6DU6
E-value=2.5928e-47,
Score=472
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
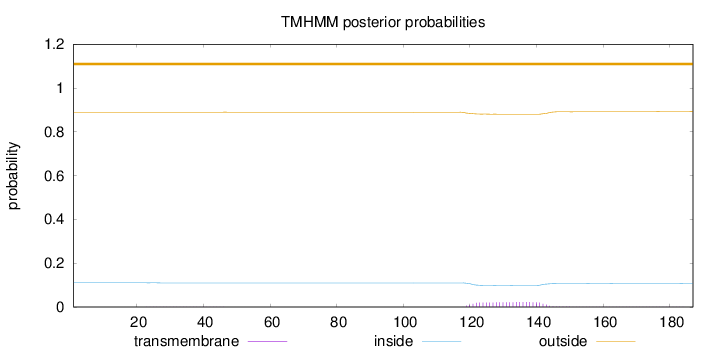
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51561
Exp number, first 60 AAs:
0.01227
Total prob of N-in:
0.11057
outside
1 - 187
Population Genetic Test Statistics
Pi
172.104067
Theta
190.829745
Tajima's D
-0.724061
CLR
0
CSRT
0.195240237988101
Interpretation
Uncertain
