Gene
KWMTBOMO04977
Pre Gene Modal
BGIBMGA004637
Annotation
PREDICTED:_protein_AAR2_homolog_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.253
Sequence
CDS
ATGGATCAAGAAACAGCGAAAAAGCTGCTAGTGGAAGGGGGCACGTTTGTATTCTTGAGAGTCCCTTCAGAAACCCAGTTTGGCATCGATATGCAGTGCTGGAACACGGAGGAAGATTTTCGTGGCGTTAAAATGATTCCTCCAGGCTTACATTATGTTCATTACTCTGCTGTCAGTAAAGGTACTGGCGATATTGCACCTAGATCTGGCTTCATGCATTACTTTCACAAAAAAGAATTTCTTGTGAAAATGTGGGACAAAAAAACTGAAGATTTGAGCAAGGAAGAGATCAGTGATGAGAGCATCCAACGTTTGAAAGATAATTTATTTAATTTGGACCGACATCTTGCTCCATATCCATATGAAATATGGCAGAAGTGGAAGTTACTGTCTTCACAGATCACTGCTGAACTAGCAGCAAAATTGGCACCAGAAAGTGGAATGATAAGAGCATCAGTTGAGCTTTTATCGATGACTGATGCAGAAAGACCAAGAGGAGTCAAGTCTTCATCTAGTTCAATAAGTCAGGAGAATGAAAAATCTCCCACTAAAGAGGATTTCAGTGAAGAACAGATTGCCGGACACTCAAGTGCTAAGAGAGCTAAAAGAATCACAAGGGAAGAGAAAGAAAAGTCATTGTTACCAGATTTGAAACCAGTTCCAGGAGCTGCTATGAGATTCACAGAAATACCTCAAGATAAATATCCACCGGATGCAACACCAGAGGAGATCACGAAACATTACTTAGATCAGTCATACTCATTAGAGCTCATGATAGCACAGCATAATGAACCTCTACACATCATAGGAGAACTGCAATTCGCATATTTGTGCTTTTTGATTGGGCACTCCCTAGAGGCTTTTGAGCACTGGAAGAGCCTCGTCATACTCATATGTTCGTGTGACGAGGCCATACACAAATACAGAAGTGTTTTCTTTCACTTCATTAGGACTATTGAGGTGCAAATCGAAGAGATGCCCGAAGATTTCTTAGCCGATATAGTGATGAATAAAAATTTAGTATACAAGAAACTCCGTGAGTTTTTCCGCACGGCTTACGTAAGCAAAGTCGACGGCAGACTTTTGACGATGATTGAACGCTTCAAAGATAACCTGACCGACAAATTGCAATGGGATTTCACTGGTCTAGACGCTGACGATGAAGACGAGAGGCCGGTCGTAGTGAAACTCGACGATGGAACTCAATAG
Protein
MDQETAKKLLVEGGTFVFLRVPSETQFGIDMQCWNTEEDFRGVKMIPPGLHYVHYSAVSKGTGDIAPRSGFMHYFHKKEFLVKMWDKKTEDLSKEEISDESIQRLKDNLFNLDRHLAPYPYEIWQKWKLLSSQITAELAAKLAPESGMIRASVELLSMTDAERPRGVKSSSSSISQENEKSPTKEDFSEEQIAGHSSAKRAKRITREEKEKSLLPDLKPVPGAAMRFTEIPQDKYPPDATPEEITKHYLDQSYSLELMIAQHNEPLHIIGELQFAYLCFLIGHSLEAFEHWKSLVILICSCDEAIHKYRSVFFHFIRTIEVQIEEMPEDFLADIVMNKNLVYKKLREFFRTAYVSKVDGRLLTMIERFKDNLTDKLQWDFTGLDADDEDERPVVVKLDDGTQ
Summary
Uniprot
H9J547
A0A1E1W634
S4PCX9
A0A212FH60
A0A2H1V6D4
A0A1E1W989
+ More
A0A0L7KUY9 A0A194QR63 A0A194PE26 A0A067RAK2 A0A2J7QZV1 A0A1W4XD50 A0A154P5C3 E2BZD7 D6W9G5 A0A0J7KL99 A0A026WWZ3 A0A1Y1KKA5 K7J1C6 E2AJX0 A0A3L8DCU9 A0A0L7QVU5 A0A151XDH3 N6U277 A0A2A3EA49 A0A088A8G6 A0A195C2P4 A0A0A9Y7M7 A0A151J8C7 A0A023F832 A0A0P4VRP8 R4FMA7 A0A0V0G9V5 A0A158P3I1 A0A1B6EZA2 A0A1B6JZV6 A0A195B676 A0A210QHS6 V3YZ02 A0A2P8Z1P5 A0A1S3JYJ5 E9HC85 F4WCT5 A0A1S3D678 A0A2L2YDZ8 A0A232FI25 A0A2L2YE17 G3NDG8 V9KWZ6 A0A3Q1GBB1 A0A3Q1GB98 A0A3Q3S218 Q6GQL8 F1R8D9 A0A3Q3FBC2 A0A2I4BTR3 A0A3S4REN9 A0A3P9PXM1 A0A3B5LEW0 M4AMA9 A0A3B5LSB8 A0A3P8SG91 A0A3Q1B8K6 A0A3B4A283 A0A087XLV6 A0A3B3VA74 A0A2S2QBK3 A0A3B5B028 A0A1A8RU85 A0A3P9JN18 A0A3P9LLJ1 T1KF33 A0A3B5BIQ7 H2M0R9 A0A3Q3DDB6 A0A1A8NHL0 A0A1A7XKJ0 A0A3Q1HS76 A0A1A8H9Y5 A0A3Q3B502 C0H960 A0A3Q3B3S5 A0A0S7GVY5 A0A3Q2CSN7 A0A3Q2E0E8 A0A1A8K2W0 A0A3Q3IJ55 A0A1A7ZS52 A0A3B4UVI7 A0A1A8DJN6 A0A1A8U6T8 A0A3B4UVJ7 A0A1A8MK67 W4YN28 A0A3B4YXU7 A0A2I0U694 J9JXH2 A0A2T7PTG8 A0A3B4EXI2 A0A315W8F0
A0A0L7KUY9 A0A194QR63 A0A194PE26 A0A067RAK2 A0A2J7QZV1 A0A1W4XD50 A0A154P5C3 E2BZD7 D6W9G5 A0A0J7KL99 A0A026WWZ3 A0A1Y1KKA5 K7J1C6 E2AJX0 A0A3L8DCU9 A0A0L7QVU5 A0A151XDH3 N6U277 A0A2A3EA49 A0A088A8G6 A0A195C2P4 A0A0A9Y7M7 A0A151J8C7 A0A023F832 A0A0P4VRP8 R4FMA7 A0A0V0G9V5 A0A158P3I1 A0A1B6EZA2 A0A1B6JZV6 A0A195B676 A0A210QHS6 V3YZ02 A0A2P8Z1P5 A0A1S3JYJ5 E9HC85 F4WCT5 A0A1S3D678 A0A2L2YDZ8 A0A232FI25 A0A2L2YE17 G3NDG8 V9KWZ6 A0A3Q1GBB1 A0A3Q1GB98 A0A3Q3S218 Q6GQL8 F1R8D9 A0A3Q3FBC2 A0A2I4BTR3 A0A3S4REN9 A0A3P9PXM1 A0A3B5LEW0 M4AMA9 A0A3B5LSB8 A0A3P8SG91 A0A3Q1B8K6 A0A3B4A283 A0A087XLV6 A0A3B3VA74 A0A2S2QBK3 A0A3B5B028 A0A1A8RU85 A0A3P9JN18 A0A3P9LLJ1 T1KF33 A0A3B5BIQ7 H2M0R9 A0A3Q3DDB6 A0A1A8NHL0 A0A1A7XKJ0 A0A3Q1HS76 A0A1A8H9Y5 A0A3Q3B502 C0H960 A0A3Q3B3S5 A0A0S7GVY5 A0A3Q2CSN7 A0A3Q2E0E8 A0A1A8K2W0 A0A3Q3IJ55 A0A1A7ZS52 A0A3B4UVI7 A0A1A8DJN6 A0A1A8U6T8 A0A3B4UVJ7 A0A1A8MK67 W4YN28 A0A3B4YXU7 A0A2I0U694 J9JXH2 A0A2T7PTG8 A0A3B4EXI2 A0A315W8F0
Pubmed
19121390
23622113
22118469
26227816
26354079
24845553
+ More
20798317 18362917 19820115 24508170 28004739 20075255 30249741 23537049 25401762 26823975 25474469 27129103 21347285 28812685 23254933 29403074 21292972 21719571 26561354 28648823 24402279 23594743 23542700 25463417 17554307 20433749 29703783
20798317 18362917 19820115 24508170 28004739 20075255 30249741 23537049 25401762 26823975 25474469 27129103 21347285 28812685 23254933 29403074 21292972 21719571 26561354 28648823 24402279 23594743 23542700 25463417 17554307 20433749 29703783
EMBL
BABH01042340
GDQN01008621
JAT82433.1
GAIX01003901
JAA88659.1
AGBW02008537
+ More
OWR53069.1 ODYU01000922 SOQ36415.1 GDQN01007518 JAT83536.1 JTDY01005377 KOB67063.1 KQ461198 KPJ06021.1 KQ459606 KPI91278.1 KK852634 KDR19779.1 NEVH01009067 PNF34112.1 KQ434820 KZC07052.1 GL451589 EFN78947.1 KQ971312 EEZ98158.1 LBMM01006019 KMQ91011.1 KK107077 EZA60567.1 GEZM01081159 JAV61839.1 GL440100 EFN66286.1 QOIP01000010 RLU17748.1 KQ414721 KOC62742.1 KQ982294 KYQ58399.1 APGK01045371 APGK01045372 KB741034 KB632397 ENN74726.1 ERL94601.1 KZ288325 PBC28056.1 KQ978379 KYM94481.1 GBHO01015993 GBRD01009596 GDHC01016977 JAG27611.1 JAG56228.1 JAQ01652.1 KQ979565 KYN21034.1 GBBI01001309 JAC17403.1 GDKW01001357 JAI55238.1 ACPB03000924 GAHY01001068 JAA76442.1 GECL01001488 JAP04636.1 ADTU01008047 ADTU01008048 ADTU01008049 GECZ01026525 JAS43244.1 GECU01002967 JAT04740.1 KQ976585 KYM79760.1 NEDP02003646 OWF48151.1 KB203711 ESO83353.1 PYGN01000239 PSN50423.1 GL732619 EFX70579.1 GL888073 EGI67980.1 IAAA01028503 LAA06358.1 NNAY01000213 OXU29977.1 IAAA01028504 LAA06359.1 JW870753 AFP03271.1 BC072724 AAH72724.1 AL954866 NCKU01000507 RWS15254.1 AYCK01007013 GGMS01005922 MBY75125.1 HAEH01005853 HAEI01009814 SBS08849.1 CAEY01000032 HAEG01002963 SBR68297.1 HADW01017221 HADX01004864 SBP18621.1 HAEB01005873 HAEC01012086 SBQ80303.1 BT058866 ACN10579.1 GBYX01449186 GBYX01449184 JAO32299.1 HAED01017331 HAEE01005949 SBR25969.1 HADY01006646 SBP45131.1 HADZ01012911 HAEA01004599 SBQ33079.1 HAEJ01002625 SBS43082.1 HAEF01015797 SBR56956.1 AAGJ04131631 KZ506099 PKU41588.1 ABLF02038670 PZQS01000002 PVD36721.1 NHOQ01000244 PWA31906.1
OWR53069.1 ODYU01000922 SOQ36415.1 GDQN01007518 JAT83536.1 JTDY01005377 KOB67063.1 KQ461198 KPJ06021.1 KQ459606 KPI91278.1 KK852634 KDR19779.1 NEVH01009067 PNF34112.1 KQ434820 KZC07052.1 GL451589 EFN78947.1 KQ971312 EEZ98158.1 LBMM01006019 KMQ91011.1 KK107077 EZA60567.1 GEZM01081159 JAV61839.1 GL440100 EFN66286.1 QOIP01000010 RLU17748.1 KQ414721 KOC62742.1 KQ982294 KYQ58399.1 APGK01045371 APGK01045372 KB741034 KB632397 ENN74726.1 ERL94601.1 KZ288325 PBC28056.1 KQ978379 KYM94481.1 GBHO01015993 GBRD01009596 GDHC01016977 JAG27611.1 JAG56228.1 JAQ01652.1 KQ979565 KYN21034.1 GBBI01001309 JAC17403.1 GDKW01001357 JAI55238.1 ACPB03000924 GAHY01001068 JAA76442.1 GECL01001488 JAP04636.1 ADTU01008047 ADTU01008048 ADTU01008049 GECZ01026525 JAS43244.1 GECU01002967 JAT04740.1 KQ976585 KYM79760.1 NEDP02003646 OWF48151.1 KB203711 ESO83353.1 PYGN01000239 PSN50423.1 GL732619 EFX70579.1 GL888073 EGI67980.1 IAAA01028503 LAA06358.1 NNAY01000213 OXU29977.1 IAAA01028504 LAA06359.1 JW870753 AFP03271.1 BC072724 AAH72724.1 AL954866 NCKU01000507 RWS15254.1 AYCK01007013 GGMS01005922 MBY75125.1 HAEH01005853 HAEI01009814 SBS08849.1 CAEY01000032 HAEG01002963 SBR68297.1 HADW01017221 HADX01004864 SBP18621.1 HAEB01005873 HAEC01012086 SBQ80303.1 BT058866 ACN10579.1 GBYX01449186 GBYX01449184 JAO32299.1 HAED01017331 HAEE01005949 SBR25969.1 HADY01006646 SBP45131.1 HADZ01012911 HAEA01004599 SBQ33079.1 HAEJ01002625 SBS43082.1 HAEF01015797 SBR56956.1 AAGJ04131631 KZ506099 PKU41588.1 ABLF02038670 PZQS01000002 PVD36721.1 NHOQ01000244 PWA31906.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000053268
UP000027135
+ More
UP000235965 UP000192223 UP000076502 UP000008237 UP000007266 UP000036403 UP000053097 UP000002358 UP000000311 UP000279307 UP000053825 UP000075809 UP000019118 UP000030742 UP000242457 UP000005203 UP000078542 UP000078492 UP000015103 UP000005205 UP000078540 UP000242188 UP000030746 UP000245037 UP000085678 UP000000305 UP000007755 UP000079169 UP000215335 UP000007635 UP000257200 UP000261640 UP000000437 UP000261660 UP000192220 UP000285301 UP000242638 UP000261380 UP000002852 UP000265080 UP000257160 UP000261520 UP000028760 UP000261500 UP000261400 UP000265200 UP000265180 UP000015104 UP000001038 UP000264820 UP000265040 UP000264800 UP000087266 UP000265020 UP000261600 UP000261420 UP000007110 UP000261360 UP000007819 UP000245119 UP000261460
UP000235965 UP000192223 UP000076502 UP000008237 UP000007266 UP000036403 UP000053097 UP000002358 UP000000311 UP000279307 UP000053825 UP000075809 UP000019118 UP000030742 UP000242457 UP000005203 UP000078542 UP000078492 UP000015103 UP000005205 UP000078540 UP000242188 UP000030746 UP000245037 UP000085678 UP000000305 UP000007755 UP000079169 UP000215335 UP000007635 UP000257200 UP000261640 UP000000437 UP000261660 UP000192220 UP000285301 UP000242638 UP000261380 UP000002852 UP000265080 UP000257160 UP000261520 UP000028760 UP000261500 UP000261400 UP000265200 UP000265180 UP000015104 UP000001038 UP000264820 UP000265040 UP000264800 UP000087266 UP000265020 UP000261600 UP000261420 UP000007110 UP000261360 UP000007819 UP000245119 UP000261460
Pfam
PF05282 AAR2
Interpro
SUPFAM
SSF51735
SSF51735
Gene 3D
ProteinModelPortal
H9J547
A0A1E1W634
S4PCX9
A0A212FH60
A0A2H1V6D4
A0A1E1W989
+ More
A0A0L7KUY9 A0A194QR63 A0A194PE26 A0A067RAK2 A0A2J7QZV1 A0A1W4XD50 A0A154P5C3 E2BZD7 D6W9G5 A0A0J7KL99 A0A026WWZ3 A0A1Y1KKA5 K7J1C6 E2AJX0 A0A3L8DCU9 A0A0L7QVU5 A0A151XDH3 N6U277 A0A2A3EA49 A0A088A8G6 A0A195C2P4 A0A0A9Y7M7 A0A151J8C7 A0A023F832 A0A0P4VRP8 R4FMA7 A0A0V0G9V5 A0A158P3I1 A0A1B6EZA2 A0A1B6JZV6 A0A195B676 A0A210QHS6 V3YZ02 A0A2P8Z1P5 A0A1S3JYJ5 E9HC85 F4WCT5 A0A1S3D678 A0A2L2YDZ8 A0A232FI25 A0A2L2YE17 G3NDG8 V9KWZ6 A0A3Q1GBB1 A0A3Q1GB98 A0A3Q3S218 Q6GQL8 F1R8D9 A0A3Q3FBC2 A0A2I4BTR3 A0A3S4REN9 A0A3P9PXM1 A0A3B5LEW0 M4AMA9 A0A3B5LSB8 A0A3P8SG91 A0A3Q1B8K6 A0A3B4A283 A0A087XLV6 A0A3B3VA74 A0A2S2QBK3 A0A3B5B028 A0A1A8RU85 A0A3P9JN18 A0A3P9LLJ1 T1KF33 A0A3B5BIQ7 H2M0R9 A0A3Q3DDB6 A0A1A8NHL0 A0A1A7XKJ0 A0A3Q1HS76 A0A1A8H9Y5 A0A3Q3B502 C0H960 A0A3Q3B3S5 A0A0S7GVY5 A0A3Q2CSN7 A0A3Q2E0E8 A0A1A8K2W0 A0A3Q3IJ55 A0A1A7ZS52 A0A3B4UVI7 A0A1A8DJN6 A0A1A8U6T8 A0A3B4UVJ7 A0A1A8MK67 W4YN28 A0A3B4YXU7 A0A2I0U694 J9JXH2 A0A2T7PTG8 A0A3B4EXI2 A0A315W8F0
A0A0L7KUY9 A0A194QR63 A0A194PE26 A0A067RAK2 A0A2J7QZV1 A0A1W4XD50 A0A154P5C3 E2BZD7 D6W9G5 A0A0J7KL99 A0A026WWZ3 A0A1Y1KKA5 K7J1C6 E2AJX0 A0A3L8DCU9 A0A0L7QVU5 A0A151XDH3 N6U277 A0A2A3EA49 A0A088A8G6 A0A195C2P4 A0A0A9Y7M7 A0A151J8C7 A0A023F832 A0A0P4VRP8 R4FMA7 A0A0V0G9V5 A0A158P3I1 A0A1B6EZA2 A0A1B6JZV6 A0A195B676 A0A210QHS6 V3YZ02 A0A2P8Z1P5 A0A1S3JYJ5 E9HC85 F4WCT5 A0A1S3D678 A0A2L2YDZ8 A0A232FI25 A0A2L2YE17 G3NDG8 V9KWZ6 A0A3Q1GBB1 A0A3Q1GB98 A0A3Q3S218 Q6GQL8 F1R8D9 A0A3Q3FBC2 A0A2I4BTR3 A0A3S4REN9 A0A3P9PXM1 A0A3B5LEW0 M4AMA9 A0A3B5LSB8 A0A3P8SG91 A0A3Q1B8K6 A0A3B4A283 A0A087XLV6 A0A3B3VA74 A0A2S2QBK3 A0A3B5B028 A0A1A8RU85 A0A3P9JN18 A0A3P9LLJ1 T1KF33 A0A3B5BIQ7 H2M0R9 A0A3Q3DDB6 A0A1A8NHL0 A0A1A7XKJ0 A0A3Q1HS76 A0A1A8H9Y5 A0A3Q3B502 C0H960 A0A3Q3B3S5 A0A0S7GVY5 A0A3Q2CSN7 A0A3Q2E0E8 A0A1A8K2W0 A0A3Q3IJ55 A0A1A7ZS52 A0A3B4UVI7 A0A1A8DJN6 A0A1A8U6T8 A0A3B4UVJ7 A0A1A8MK67 W4YN28 A0A3B4YXU7 A0A2I0U694 J9JXH2 A0A2T7PTG8 A0A3B4EXI2 A0A315W8F0
Ontologies
GO
PANTHER
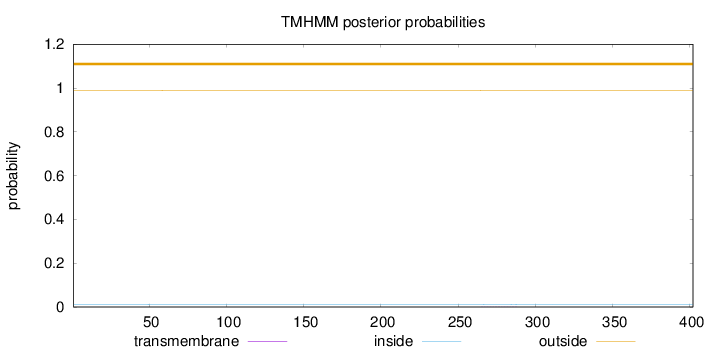
Topology
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01969
Exp number, first 60 AAs:
0.00209
Total prob of N-in:
0.01209
outside
1 - 402
Population Genetic Test Statistics
Pi
282.177362
Theta
184.299245
Tajima's D
1.860307
CLR
0
CSRT
0.859807009649518
Interpretation
Uncertain
