Gene
KWMTBOMO04971
Pre Gene Modal
BGIBMGA008087
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_DDX11_[Papilio_xuthus]
Full name
ATP-dependent DNA helicase DDX11
Alternative Name
DEAD/H-box protein 11
CHL1-related protein 1
Keratinocyte growth factor-regulated gene 2 protein
CHL1-related protein 1
Keratinocyte growth factor-regulated gene 2 protein
Location in the cell
Nuclear Reliability : 3.73
Sequence
CDS
ATGGAGCCAGTATTTCAATTTCCTTTTAATCCATATCCCATTCAAGAAAAGTTCATGCGCGAGCTTTATTTTACCATAGAAAATAGAAAATTGGGAATATTCGAAAGTCCAACTGGAACAGGTAAATCACTTAGCATATGTTGTGCAGCGCTCCAGTGGCTTAAAGATAGCAATAAGAAAATTTTAGAAGAACTAAAAGCTGAAATTGGCACTCTTAAATTGGAAATTTCACAAACTATGGACAGTAATGATTGGCTGCAAGAAGAATATGAAAAAATTAAGAAAAACCAAATGCTCATAACACTACAAAATAAAATGGATAAAATTATACAATGTAAAGAAAATTTTGAGAATATGGCAAAGCGAGTTTCAAAACATAAATCAAATGTTAACGTTAAGTCAACAGATAACTATTGTAAGAGTGATAAGAAAAAAAACAGTGAACCTGAATTAGAGAAAGAGAACATAGAAGAACAGAATGATAGTGAAGGTGATAGAGATTTAATTCTTGAGGAATTTGAAGAAAATGCTGAAAGTGATACTTATCCTGAGGAAGTGGAAGAAATGGACAATAAAGATGAAATCATAAAGGTATACATTAGCAGCAGGACACATAGTCAGCTCTCGCAGTTTATTGGGGAAATAAAGAGAACCATTTTCAAGGATGTCACTAGAGTTGTCACTTTAGCATCGAGACAACATTACTGCATCAATTCTGAAGTCGCAAAGTTGAAGAATGTTAATTTAATAAACGAAAGGTGTCTAGATATGCAGAAAACTAAAAGTAAATCAACATCATTCAGTGACGATGGTAGAGTTCTCAAAAAGAGTAAGTTAAGGGCTTGTACTGCCTGCCCTTATTACAATCAAAACAATATAACAAAACTAAAAGAGAGAATGCTAATTGAAATTATGGACATGGAAGACTTGGTCAAATGTGGAAAAGAATTGAAGGCTTGTCCATATTATGCGTCTAGGATGGCCATGGATGATGCAGAGGTGGTACTTATAAGTCACGCGGGTATAGTAAGTTCTGGAGCCAGATCAGGAATATCATTAAAATTGAAGGATAACATCTTGATTCTGGATGAAGCACATGGCCTCAGTGCGGCACTGGAGAATGCTCATTCCGCCCCTGTCACTGTTAAACAGCTGTCTTCCGTGAAAACTTATCTACAATTCTACATAAACAAGTATAGAGCAAGATTGAGTAGTAGAAATTTGTTGTCTCTGAACCAGATTAGTTTTGTTGTAGGGAAGTTAATAAGCTTAGTTAAACCAAAAGAGACAGAAAAAAGCAAAGAAACGACCATACTTACTCTTGAGGATTTCGTGATAAAAGCTGAAATAGATCACATGAATTTGAGGCCCTTAGTAGAATTTTGTAGAAGCACACGTTTGGCGCCCAAATTACACGGGTTTTCCATGAGGTATAGCCAGCAAGTATTGGAGGAGGAGGAATCAAAAAAAATTGCAGTGAAAAAAAACTCATTCCAGAAATTTCTATCAAATATCGCAAAGAAGAAGGAATTGGCTAGCGTTGAAGAAACACAAACGGAAACTAAACCTATCGTCCAGTTCGCAACAGAAACGTCGGCGGGCAGCGGGCTCTATGCCGTGCTAGACTTCCTGGAAATGCTCTGCGCACGAAGCGAGAACGGACGAGTGCTGGTGCAACAAGGCGACACCGGCCTGCTCAAGTATCTGCTGCTCAATCCGGCTGAATGTTTTACTGATGTACTGGCTCAATGCAGATCTGTGATTGTGGCTGGGGGTACCATGGAGCCGATAAGTGAATTCAAAGCGCTGCTCAGCTCCAACGGAAGATCAGACTCCAAACTGAACATAGTGCGATGTGAACATGTTGTACCTCCAGACAATATATTAGCTGTGTGCTTGTCAAGAGGACCGTCTAATACTGTATTCAATTTCTCCTATGAAAATCGAATGTCCAAGGAATTGCTAAATGAAGTAGGTCGTACATTAAGAAACGTCTGCAGCATCGTGCCGGGCGGTGTGGTATGCTTCCTACCTTCATATTCCTATGAACAAACTGTCTACGATCATTTGAAAAACAATAATATTATTGATTTTATAAGCAAAAGGAAGAGCGTATTCCGTGAACCAAAATCTGCAGCTGATGTTGAACAAGTACTTCACAAATATTCATTAGCTGTAAAAAGTAAGGAAAAAAATGGAGCTTTATTACTGAGCGTCGTAGGTGGTAAGCTTAGCGAAGGCTTGAATTTTAGCGACGATCTGGGGCGATGTGTAATGGTTGTGGGAATGCCGTATCCTAATGTGAAGTCGCCAGAGCTGCAGGAGAAAATGTCCTATTTGAATAGAACTGCAGGTGGTAATGCTGCTAACATTTACTATGAAAGTTTGTGTATGAAGGCAGTCAACCAATGTATAGGTAGGGCTGTTCGGCATATTAATGACTATGCAACTGTGCTGTTAATAGATGAAAGATATTCGAGAGTCCAAACGATATCAGCTTTACCTTCATTTGTACAGAAATCGTTAATATCAAACTCCAGTTTTGGTCAAATGATCGGAAGCATGGCAAAGTTCTTTTCAAGGCACAAGGAAATAAAAACAAATTGA
Protein
MEPVFQFPFNPYPIQEKFMRELYFTIENRKLGIFESPTGTGKSLSICCAALQWLKDSNKKILEELKAEIGTLKLEISQTMDSNDWLQEEYEKIKKNQMLITLQNKMDKIIQCKENFENMAKRVSKHKSNVNVKSTDNYCKSDKKKNSEPELEKENIEEQNDSEGDRDLILEEFEENAESDTYPEEVEEMDNKDEIIKVYISSRTHSQLSQFIGEIKRTIFKDVTRVVTLASRQHYCINSEVAKLKNVNLINERCLDMQKTKSKSTSFSDDGRVLKKSKLRACTACPYYNQNNITKLKERMLIEIMDMEDLVKCGKELKACPYYASRMAMDDAEVVLISHAGIVSSGARSGISLKLKDNILILDEAHGLSAALENAHSAPVTVKQLSSVKTYLQFYINKYRARLSSRNLLSLNQISFVVGKLISLVKPKETEKSKETTILTLEDFVIKAEIDHMNLRPLVEFCRSTRLAPKLHGFSMRYSQQVLEEEESKKIAVKKNSFQKFLSNIAKKKELASVEETQTETKPIVQFATETSAGSGLYAVLDFLEMLCARSENGRVLVQQGDTGLLKYLLLNPAECFTDVLAQCRSVIVAGGTMEPISEFKALLSSNGRSDSKLNIVRCEHVVPPDNILAVCLSRGPSNTVFNFSYENRMSKELLNEVGRTLRNVCSIVPGGVVCFLPSYSYEQTVYDHLKNNNIIDFISKRKSVFREPKSAADVEQVLHKYSLAVKSKEKNGALLLSVVGGKLSEGLNFSDDLGRCVMVVGMPYPNVKSPELQEKMSYLNRTAGGNAANIYYESLCMKAVNQCIGRAVRHINDYATVLLIDERYSRVQTISALPSFVQKSLISNSSFGQMIGSMAKFFSRHKEIKTN
Summary
Description
DNA-dependent ATPase and ATP-dependent DNA helicase that participates in various functions in genomic stability, including DNA replication, DNA repair and heterochromatin organization as well as in ribosomal RNA synthesis. Plays a role in DNA double-strand break (DSB) repair at the DNA replication fork during DNA replication recovery from DNA damage. Plays a role in the regulation of sister chromatid cohesion and mitotic chromosome segregation. Stimulates 5'-single-stranded DNA flap endonuclease activity of FEN1 in an ATP- and helicase-independent manner. Plays also a role in heterochromatin organization (By similarity). Involved in rRNA transcription activation through binding to active hypomethylated rDNA gene loci by recruiting UBTF and the RNA polymerase Pol I transcriptional machinery (PubMed:26089203). Plays a role in embryonic development (PubMed:26089203). Associates with chromatin at DNA replication fork regions. Binds to single- and double-stranded DNAs (By similarity).
DNA-dependent ATPase and ATP-dependent DNA helicase that participates in various functions in genomic stability, including DNA replication, DNA repair and heterochromatin organization as well as in ribosomal RNA synthesis (PubMed:10648783, PubMed:21854770, PubMed:23797032, PubMed:26089203, PubMed:26503245). Its double-stranded DNA helicase activity requires either a minimal 5'-single-stranded tail length of approximately 15 nt (flap substrates) or 10 nt length single-stranded gapped DNA substrates of a partial duplex DNA structure for helicase loading and translocation along DNA in a 5' to 3' direction (PubMed:18499658, PubMed:22102414). The helicase activity is capable of displacing duplex regions up to 100 bp, which can be extended up to 500 bp by the replication protein A (RPA) or the cohesion CTF18-replication factor C (Ctf18-RFC) complex activities (PubMed:18499658). Shows also ATPase- and helicase activities on substrates that mimic key DNA intermediates of replication, repair and homologous recombination reactions, including forked duplex, anti-parallel G-quadruplex and three-stranded D-loop DNA molecules (PubMed:22102414, PubMed:26503245). Plays a role in DNA double-strand break (DSB) repair at the DNA replication fork during DNA replication recovery from DNA damage (PubMed:23797032). Recruited with TIMELESS factor upon DNA-replication stress response at DNA replication fork to preserve replication fork progression, and hence ensure DNA replication fidelity (PubMed:26503245). Cooperates also with TIMELESS factor during DNA replication to regulate proper sister chromatid cohesion and mitotic chromosome segregation (PubMed:17105772, PubMed:18499658, PubMed:20124417, PubMed:23116066, PubMed:23797032). Stimulates 5'-single-stranded DNA flap endonuclease activity of FEN1 in an ATP- and helicase-independent manner; and hence it may contribute in Okazaki fragment processing at DNA replication fork during lagging strand DNA synthesis (PubMed:18499658). Its ability to function at DNA replication fork is modulated by its binding to long non-coding RNA (lncRNA) cohesion regulator non-coding RNA DDX11-AS1/CONCR, which is able to increase both DDX11 ATPase activity and binding to DNA replicating regions (PubMed:27477908). Plays also a role in heterochromatin organization (PubMed:21854770). Involved in rRNA transcription activation through binding to active hypomethylated rDNA gene loci by recruiting UBTF and the RNA polymerase Pol I transcriptional machinery (PubMed:26089203). Plays a role in embryonic development and prevention of aneuploidy (By similarity). Involved in melanoma cell proliferation and survival (PubMed:23116066). Associates with chromatin at DNA replication fork regions (PubMed:27477908). Binds to single- and double-stranded DNAs (PubMed:9013641, PubMed:18499658, PubMed:22102414).
(Microbial infection) Required for bovine papillomavirus type 1 regulatory protein E2 loading onto mitotic chromosomes during DNA replication for the viral genome to be maintained and segregated.
DNA-dependent ATPase and ATP-dependent DNA helicase that participates in various functions in genomic stability, including DNA replication, DNA repair and heterochromatin organization as well as in ribosomal RNA synthesis (PubMed:10648783, PubMed:21854770, PubMed:23797032, PubMed:26089203, PubMed:26503245). Its double-stranded DNA helicase activity requires either a minimal 5'-single-stranded tail length of approximately 15 nt (flap substrates) or 10 nt length single-stranded gapped DNA substrates of a partial duplex DNA structure for helicase loading and translocation along DNA in a 5' to 3' direction (PubMed:18499658, PubMed:22102414). The helicase activity is capable of displacing duplex regions up to 100 bp, which can be extended up to 500 bp by the replication protein A (RPA) or the cohesion CTF18-replication factor C (Ctf18-RFC) complex activities (PubMed:18499658). Shows also ATPase- and helicase activities on substrates that mimic key DNA intermediates of replication, repair and homologous recombination reactions, including forked duplex, anti-parallel G-quadruplex and three-stranded D-loop DNA molecules (PubMed:22102414, PubMed:26503245). Plays a role in DNA double-strand break (DSB) repair at the DNA replication fork during DNA replication recovery from DNA damage (PubMed:23797032). Recruited with TIMELESS factor upon DNA-replication stress response at DNA replication fork to preserve replication fork progression, and hence ensure DNA replication fidelity (PubMed:26503245). Cooperates also with TIMELESS factor during DNA replication to regulate proper sister chromatid cohesion and mitotic chromosome segregation (PubMed:17105772, PubMed:18499658, PubMed:20124417, PubMed:23116066, PubMed:23797032). Stimulates 5'-single-stranded DNA flap endonuclease activity of FEN1 in an ATP- and helicase-independent manner; and hence it may contribute in Okazaki fragment processing at DNA replication fork during lagging strand DNA synthesis (PubMed:18499658). Its ability to function at DNA replication fork is modulated by its binding to long non-coding RNA (lncRNA) cohesion regulator non-coding RNA DDX11-AS1/CONCR, which is able to increase both DDX11 ATPase activity and binding to DNA replicating regions (PubMed:27477908). Plays also a role in heterochromatin organization (PubMed:21854770). Involved in rRNA transcription activation through binding to active hypomethylated rDNA gene loci by recruiting UBTF and the RNA polymerase Pol I transcriptional machinery (PubMed:26089203). Plays a role in embryonic development and prevention of aneuploidy (By similarity). Involved in melanoma cell proliferation and survival (PubMed:23116066). Associates with chromatin at DNA replication fork regions (PubMed:27477908). Binds to single- and double-stranded DNAs (PubMed:9013641, PubMed:18499658, PubMed:22102414).
(Microbial infection) Required for bovine papillomavirus type 1 regulatory protein E2 loading onto mitotic chromosomes during DNA replication for the viral genome to be maintained and segregated.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
[4Fe-4S] cluster
Subunit
Associates with the CTF18-RFC complex (PubMed:18499658). Associates with a cohesin complex composed of RAD21, SMC1 proteins and SMC3 (PubMed:17105772). Interacts with CHTF18 (PubMed:18499658). Interacts with DSCC1 (PubMed:18499658). Interacts with FEN1; this interaction is direct and increases flap endonuclease activity of FEN1 (PubMed:18499658). Interacts with PCNA (PubMed:18499658). Interacts with POLR1A and UBTF (PubMed:26089203). Interacts with RAD21, SMC1 proteins and SMC3 (PubMed:17105772). Interacts with RFC2 (PubMed:18499658). Interacts with TIMELESS; this interaction increases recruitment of both proteins onto chromatin in response to replication stress induction by hydroxyurea (PubMed:20124417, PubMed:26503245).
(Microbial infection) Interacts with bovine papillomavirus type 1 regulatory protein E2; this interaction stimulates the recruitment of E2 onto mitotic chromosomes.
(Microbial infection) Interacts with bovine papillomavirus type 1 regulatory protein E2; this interaction stimulates the recruitment of E2 onto mitotic chromosomes.
Similarity
Belongs to the DEAD box helicase family. DEAH subfamily. DDX11/CHL1 sub-subfamily.
Keywords
4Fe-4S
Activator
ATP-binding
Complete proteome
Cytoplasm
Cytoskeleton
Developmental protein
DNA damage
DNA repair
DNA replication
DNA-binding
Helicase
Hydrolase
Iron
Iron-sulfur
Metal-binding
Nucleotide-binding
Nucleus
Reference proteome
RNA-binding
Transcription
Transcription regulation
Alternative splicing
Chromosome
Disease mutation
Host-virus interaction
Phosphoprotein
Polymorphism
Feature
chain ATP-dependent DNA helicase DDX11
splice variant In isoform 3.
sequence variant In WBRS; impairs the helicase activity by perturbing its DNA binding and DNA-dependent ATPase activity; reduces binding to rDNA promoter and promotion of rDNA transcription; dbSNP:rs201968272.
splice variant In isoform 3.
sequence variant In WBRS; impairs the helicase activity by perturbing its DNA binding and DNA-dependent ATPase activity; reduces binding to rDNA promoter and promotion of rDNA transcription; dbSNP:rs201968272.
Uniprot
H9JEZ0
A0A2H1W4V9
A0A2A4J8Q3
A0A2A4IVS0
A0A2W1BM24
A0A194PF05
+ More
A0A194QR57 A0A212F1R3 A0A067QR04 A0A1B6DBS7 A0A2J7QJI9 A0A2J7QJK9 J9K2V1 A0A1Y1N091 A0A2S2QHC2 A0A146MCY6 A0A146L2S9 A0A0A9Z2M7 K7J4C8 A0A232EV86 A0A2H8TU99 A0A023F2L6 D2A079 F4W8N7 A0A2C9L2J2 U4U4U2 N6TUW0 A0A2A3ED27 E0W3H5 A0A158NKD7 A0A3L8DE61 F1R345 B5DEB4 A0A1Q3EVL6 E2A2U0 F6WA03 E2C9U9 G1K869 A0A1W5AEN6 A0A1B0D1Q6 A0A3Q2UC34 A0A2R8QL47 R7UDW5 A0A1L8GWC6 A0A1X7UKI4 A0A2K5QEH2 A0A336MV21 A0A3Q1AUT7 A0A3B4UG89 F6US01 K1S021 A0A195B0I8 A0A099ZTU7 A0A0J7KXG7 A0A2P6KV16 Q96FC9-2 A0A336KTI8 V9KEK7 B0W2A2 A0A3B3RCP9 Q16VU1 A0A2J8TC64 K7BT75 G1QRI3 A0A210Q9K9 A0A151IJU5 A0A2I4ATX3 A0A195EIP0 A0A226PK30 A0A2I4ATZ3 A0A2I3H228 K7CE42 U3JMF8 F6RTU7 A0A2K5JVB3 G7PJR9 Q2NKM7 A0A2K6AX93 A0A0D9RAP8 A0A2K5V8E1 A0A096N2K6 A0A3N0YS60 K7AUW0 A0A2J8TC84 A0A182GLP3 H9YYA7 G7N5Y1 A0A3B4XZ05 A0A3Q7MKZ8 A0A182HL54 A0A2K5ZSF2 A0A3Q3QH78 A0A151WT71 F6WMZ3 A0A2Y9H9R3 A0A1D2MKG0 Q7PXR8 A0A2U3ZSE8 A0A3Q3LNF1 A0A2K5JVB7
A0A194QR57 A0A212F1R3 A0A067QR04 A0A1B6DBS7 A0A2J7QJI9 A0A2J7QJK9 J9K2V1 A0A1Y1N091 A0A2S2QHC2 A0A146MCY6 A0A146L2S9 A0A0A9Z2M7 K7J4C8 A0A232EV86 A0A2H8TU99 A0A023F2L6 D2A079 F4W8N7 A0A2C9L2J2 U4U4U2 N6TUW0 A0A2A3ED27 E0W3H5 A0A158NKD7 A0A3L8DE61 F1R345 B5DEB4 A0A1Q3EVL6 E2A2U0 F6WA03 E2C9U9 G1K869 A0A1W5AEN6 A0A1B0D1Q6 A0A3Q2UC34 A0A2R8QL47 R7UDW5 A0A1L8GWC6 A0A1X7UKI4 A0A2K5QEH2 A0A336MV21 A0A3Q1AUT7 A0A3B4UG89 F6US01 K1S021 A0A195B0I8 A0A099ZTU7 A0A0J7KXG7 A0A2P6KV16 Q96FC9-2 A0A336KTI8 V9KEK7 B0W2A2 A0A3B3RCP9 Q16VU1 A0A2J8TC64 K7BT75 G1QRI3 A0A210Q9K9 A0A151IJU5 A0A2I4ATX3 A0A195EIP0 A0A226PK30 A0A2I4ATZ3 A0A2I3H228 K7CE42 U3JMF8 F6RTU7 A0A2K5JVB3 G7PJR9 Q2NKM7 A0A2K6AX93 A0A0D9RAP8 A0A2K5V8E1 A0A096N2K6 A0A3N0YS60 K7AUW0 A0A2J8TC84 A0A182GLP3 H9YYA7 G7N5Y1 A0A3B4XZ05 A0A3Q7MKZ8 A0A182HL54 A0A2K5ZSF2 A0A3Q3QH78 A0A151WT71 F6WMZ3 A0A2Y9H9R3 A0A1D2MKG0 Q7PXR8 A0A2U3ZSE8 A0A3Q3LNF1 A0A2K5JVB7
EC Number
3.6.4.12
Pubmed
19121390
28756777
26354079
22118469
24845553
28004739
+ More
26823975 25401762 20075255 28648823 25474469 18362917 19820115 21719571 15562597 23537049 20566863 21347285 30249741 23594743 26089203 20798317 20431018 15592404 23254933 27762356 22992520 8798685 9013641 15489334 10648783 17105772 17189189 18499658 18669648 20137776 20124417 21269460 21854770 22102414 23116066 23797032 23186163 27477908 26503245 23033317 24402279 29240929 17510324 28812685 24621616 19892987 22002653 26483478 25319552 17431167 27289101 12364791 14747013 17210077
26823975 25401762 20075255 28648823 25474469 18362917 19820115 21719571 15562597 23537049 20566863 21347285 30249741 23594743 26089203 20798317 20431018 15592404 23254933 27762356 22992520 8798685 9013641 15489334 10648783 17105772 17189189 18499658 18669648 20137776 20124417 21269460 21854770 22102414 23116066 23797032 23186163 27477908 26503245 23033317 24402279 29240929 17510324 28812685 24621616 19892987 22002653 26483478 25319552 17431167 27289101 12364791 14747013 17210077
EMBL
BABH01023645
BABH01023646
BABH01023647
BABH01023648
BABH01023649
ODYU01006040
+ More
SOQ47534.1 NWSH01002634 PCG67792.1 NWSH01006780 PCG63240.1 KZ149974 PZC75978.1 KQ459606 KPI91284.1 KQ461198 KPJ06016.1 AGBW02010825 OWR47680.1 KK853067 KDR11841.1 GEDC01014155 GEDC01002826 JAS23143.1 JAS34472.1 NEVH01013555 PNF28755.1 PNF28756.1 ABLF02040828 GEZM01016229 JAV91319.1 GGMS01007369 MBY76572.1 GDHC01000981 JAQ17648.1 GDHC01017122 JAQ01507.1 GBHO01007539 GDHC01006020 JAG36065.1 JAQ12609.1 AAZX01007303 NNAY01002030 OXU22266.1 GFXV01006020 MBW17825.1 GBBI01003438 JAC15274.1 KQ971338 EFA02893.2 GL887908 EGI69528.1 KB631729 ERL85626.1 APGK01017607 KB740045 ENN81863.1 KZ288279 PBC29683.1 DS235882 EEB20181.1 ADTU01018786 QOIP01000009 RLU18770.1 CU928088 BC085645 BC168601 AAI68601.1 GFDL01015702 JAV19343.1 GL436214 EFN72247.1 AAMC01120252 AAMC01120253 AAMC01120254 AAMC01120255 AAMC01120256 GL453902 EFN75286.1 AJVK01022248 AC174384 AMQN01008211 KB302448 ELU04301.1 CM004470 OCT88134.1 UFQT01003028 SSX34454.1 JH817532 EKC40591.1 KQ976692 KYM77805.1 KL897454 KGL84533.1 LBMM01002329 KMQ94974.1 MWRG01004872 PRD30168.1 X99583 U33833 U75967 U75968 BC050069 BC050522 UFQS01000768 UFQT01000768 SSX06735.1 SSX27080.1 JW863672 AFO96189.1 DS231826 EDS28717.1 CH477581 EAT38683.1 NDHI03003510 PNJ30627.1 GABF01004861 GABD01003735 JAA17284.1 JAA29365.1 ADFV01028735 ADFV01028736 NEDP02004513 OWF45416.1 KQ977294 KYN03917.1 KQ978881 KYN27729.1 AWGT02000049 OXB80435.1 GABE01003875 GABE01003874 GABE01003873 JAA40866.1 AGTO01001282 CM001286 EHH66054.1 BC111733 AAI11734.1 AQIB01151485 AQIA01009999 AHZZ02001665 RJVU01027559 ROL49004.1 GABC01007763 JAA03575.1 PNJ30640.1 JXUM01072556 JXUM01072557 KQ562727 KXJ75242.1 JU471869 AFH28673.1 CM001263 EHH20478.1 APCN01000881 KQ982762 KYQ51038.1 JSUE03006122 LJIJ01000997 ODM93421.1 AAAB01008987 EAA01808.4
SOQ47534.1 NWSH01002634 PCG67792.1 NWSH01006780 PCG63240.1 KZ149974 PZC75978.1 KQ459606 KPI91284.1 KQ461198 KPJ06016.1 AGBW02010825 OWR47680.1 KK853067 KDR11841.1 GEDC01014155 GEDC01002826 JAS23143.1 JAS34472.1 NEVH01013555 PNF28755.1 PNF28756.1 ABLF02040828 GEZM01016229 JAV91319.1 GGMS01007369 MBY76572.1 GDHC01000981 JAQ17648.1 GDHC01017122 JAQ01507.1 GBHO01007539 GDHC01006020 JAG36065.1 JAQ12609.1 AAZX01007303 NNAY01002030 OXU22266.1 GFXV01006020 MBW17825.1 GBBI01003438 JAC15274.1 KQ971338 EFA02893.2 GL887908 EGI69528.1 KB631729 ERL85626.1 APGK01017607 KB740045 ENN81863.1 KZ288279 PBC29683.1 DS235882 EEB20181.1 ADTU01018786 QOIP01000009 RLU18770.1 CU928088 BC085645 BC168601 AAI68601.1 GFDL01015702 JAV19343.1 GL436214 EFN72247.1 AAMC01120252 AAMC01120253 AAMC01120254 AAMC01120255 AAMC01120256 GL453902 EFN75286.1 AJVK01022248 AC174384 AMQN01008211 KB302448 ELU04301.1 CM004470 OCT88134.1 UFQT01003028 SSX34454.1 JH817532 EKC40591.1 KQ976692 KYM77805.1 KL897454 KGL84533.1 LBMM01002329 KMQ94974.1 MWRG01004872 PRD30168.1 X99583 U33833 U75967 U75968 BC050069 BC050522 UFQS01000768 UFQT01000768 SSX06735.1 SSX27080.1 JW863672 AFO96189.1 DS231826 EDS28717.1 CH477581 EAT38683.1 NDHI03003510 PNJ30627.1 GABF01004861 GABD01003735 JAA17284.1 JAA29365.1 ADFV01028735 ADFV01028736 NEDP02004513 OWF45416.1 KQ977294 KYN03917.1 KQ978881 KYN27729.1 AWGT02000049 OXB80435.1 GABE01003875 GABE01003874 GABE01003873 JAA40866.1 AGTO01001282 CM001286 EHH66054.1 BC111733 AAI11734.1 AQIB01151485 AQIA01009999 AHZZ02001665 RJVU01027559 ROL49004.1 GABC01007763 JAA03575.1 PNJ30640.1 JXUM01072556 JXUM01072557 KQ562727 KXJ75242.1 JU471869 AFH28673.1 CM001263 EHH20478.1 APCN01000881 KQ982762 KYQ51038.1 JSUE03006122 LJIJ01000997 ODM93421.1 AAAB01008987 EAA01808.4
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000235965 UP000007819 UP000002358 UP000215335 UP000007266 UP000007755 UP000076420 UP000030742 UP000019118 UP000242457 UP000009046 UP000005205 UP000279307 UP000000437 UP000000311 UP000008143 UP000008237 UP000001646 UP000192224 UP000092462 UP000000539 UP000014760 UP000186698 UP000007879 UP000233040 UP000257160 UP000261420 UP000005408 UP000078540 UP000053641 UP000036403 UP000005640 UP000002320 UP000261540 UP000008820 UP000001073 UP000242188 UP000078542 UP000192220 UP000078492 UP000198419 UP000016665 UP000002281 UP000233080 UP000009130 UP000233120 UP000029965 UP000233100 UP000028761 UP000069940 UP000249989 UP000261360 UP000286641 UP000075840 UP000233140 UP000261600 UP000075809 UP000006718 UP000248481 UP000094527 UP000007062 UP000245340 UP000261640
UP000235965 UP000007819 UP000002358 UP000215335 UP000007266 UP000007755 UP000076420 UP000030742 UP000019118 UP000242457 UP000009046 UP000005205 UP000279307 UP000000437 UP000000311 UP000008143 UP000008237 UP000001646 UP000192224 UP000092462 UP000000539 UP000014760 UP000186698 UP000007879 UP000233040 UP000257160 UP000261420 UP000005408 UP000078540 UP000053641 UP000036403 UP000005640 UP000002320 UP000261540 UP000008820 UP000001073 UP000242188 UP000078542 UP000192220 UP000078492 UP000198419 UP000016665 UP000002281 UP000233080 UP000009130 UP000233120 UP000029965 UP000233100 UP000028761 UP000069940 UP000249989 UP000261360 UP000286641 UP000075840 UP000233140 UP000261600 UP000075809 UP000006718 UP000248481 UP000094527 UP000007062 UP000245340 UP000261640
Interpro
IPR014013
Helic_SF1/SF2_ATP-bd_DinG/Rad3
+ More
IPR006554 Helicase-like_DEXD_c2
IPR013020 Rad3/Chl1-like
IPR006555 ATP-dep_Helicase_C
IPR010614 DEAD_2
IPR028331 CHL1/DDX11
IPR027417 P-loop_NTPase
IPR014001 Helicase_ATP-bd
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR001910 Inosine/uridine_hydrolase_dom
IPR036452 Ribo_hydro-like
IPR026321 Coiled-coil_dom_con_pro_134
IPR006554 Helicase-like_DEXD_c2
IPR013020 Rad3/Chl1-like
IPR006555 ATP-dep_Helicase_C
IPR010614 DEAD_2
IPR028331 CHL1/DDX11
IPR027417 P-loop_NTPase
IPR014001 Helicase_ATP-bd
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR001910 Inosine/uridine_hydrolase_dom
IPR036452 Ribo_hydro-like
IPR026321 Coiled-coil_dom_con_pro_134
Gene 3D
ProteinModelPortal
H9JEZ0
A0A2H1W4V9
A0A2A4J8Q3
A0A2A4IVS0
A0A2W1BM24
A0A194PF05
+ More
A0A194QR57 A0A212F1R3 A0A067QR04 A0A1B6DBS7 A0A2J7QJI9 A0A2J7QJK9 J9K2V1 A0A1Y1N091 A0A2S2QHC2 A0A146MCY6 A0A146L2S9 A0A0A9Z2M7 K7J4C8 A0A232EV86 A0A2H8TU99 A0A023F2L6 D2A079 F4W8N7 A0A2C9L2J2 U4U4U2 N6TUW0 A0A2A3ED27 E0W3H5 A0A158NKD7 A0A3L8DE61 F1R345 B5DEB4 A0A1Q3EVL6 E2A2U0 F6WA03 E2C9U9 G1K869 A0A1W5AEN6 A0A1B0D1Q6 A0A3Q2UC34 A0A2R8QL47 R7UDW5 A0A1L8GWC6 A0A1X7UKI4 A0A2K5QEH2 A0A336MV21 A0A3Q1AUT7 A0A3B4UG89 F6US01 K1S021 A0A195B0I8 A0A099ZTU7 A0A0J7KXG7 A0A2P6KV16 Q96FC9-2 A0A336KTI8 V9KEK7 B0W2A2 A0A3B3RCP9 Q16VU1 A0A2J8TC64 K7BT75 G1QRI3 A0A210Q9K9 A0A151IJU5 A0A2I4ATX3 A0A195EIP0 A0A226PK30 A0A2I4ATZ3 A0A2I3H228 K7CE42 U3JMF8 F6RTU7 A0A2K5JVB3 G7PJR9 Q2NKM7 A0A2K6AX93 A0A0D9RAP8 A0A2K5V8E1 A0A096N2K6 A0A3N0YS60 K7AUW0 A0A2J8TC84 A0A182GLP3 H9YYA7 G7N5Y1 A0A3B4XZ05 A0A3Q7MKZ8 A0A182HL54 A0A2K5ZSF2 A0A3Q3QH78 A0A151WT71 F6WMZ3 A0A2Y9H9R3 A0A1D2MKG0 Q7PXR8 A0A2U3ZSE8 A0A3Q3LNF1 A0A2K5JVB7
A0A194QR57 A0A212F1R3 A0A067QR04 A0A1B6DBS7 A0A2J7QJI9 A0A2J7QJK9 J9K2V1 A0A1Y1N091 A0A2S2QHC2 A0A146MCY6 A0A146L2S9 A0A0A9Z2M7 K7J4C8 A0A232EV86 A0A2H8TU99 A0A023F2L6 D2A079 F4W8N7 A0A2C9L2J2 U4U4U2 N6TUW0 A0A2A3ED27 E0W3H5 A0A158NKD7 A0A3L8DE61 F1R345 B5DEB4 A0A1Q3EVL6 E2A2U0 F6WA03 E2C9U9 G1K869 A0A1W5AEN6 A0A1B0D1Q6 A0A3Q2UC34 A0A2R8QL47 R7UDW5 A0A1L8GWC6 A0A1X7UKI4 A0A2K5QEH2 A0A336MV21 A0A3Q1AUT7 A0A3B4UG89 F6US01 K1S021 A0A195B0I8 A0A099ZTU7 A0A0J7KXG7 A0A2P6KV16 Q96FC9-2 A0A336KTI8 V9KEK7 B0W2A2 A0A3B3RCP9 Q16VU1 A0A2J8TC64 K7BT75 G1QRI3 A0A210Q9K9 A0A151IJU5 A0A2I4ATX3 A0A195EIP0 A0A226PK30 A0A2I4ATZ3 A0A2I3H228 K7CE42 U3JMF8 F6RTU7 A0A2K5JVB3 G7PJR9 Q2NKM7 A0A2K6AX93 A0A0D9RAP8 A0A2K5V8E1 A0A096N2K6 A0A3N0YS60 K7AUW0 A0A2J8TC84 A0A182GLP3 H9YYA7 G7N5Y1 A0A3B4XZ05 A0A3Q7MKZ8 A0A182HL54 A0A2K5ZSF2 A0A3Q3QH78 A0A151WT71 F6WMZ3 A0A2Y9H9R3 A0A1D2MKG0 Q7PXR8 A0A2U3ZSE8 A0A3Q3LNF1 A0A2K5JVB7
PDB
6NMI
E-value=2.42853e-26,
Score=299
Ontologies
GO
GO:0005634
GO:0003677
GO:0004003
GO:0006139
GO:0005524
GO:0034085
GO:0032508
GO:0004386
GO:0006281
GO:0003723
GO:0051539
GO:0030496
GO:0007275
GO:0005737
GO:0005815
GO:1901836
GO:1990700
GO:0046872
GO:0009303
GO:0000922
GO:0005730
GO:0006260
GO:0031390
GO:0072711
GO:0006974
GO:2000781
GO:0032079
GO:0003688
GO:0008186
GO:0051880
GO:0003682
GO:0005813
GO:1904976
GO:1901838
GO:0003697
GO:0031297
GO:0035563
GO:0001650
GO:0000790
GO:0003727
GO:0045876
GO:0044806
GO:0072719
GO:0032091
GO:0045142
GO:0005654
GO:0008094
GO:0008026
GO:0007062
GO:0003690
GO:0016032
GO:0036498
GO:0070062
GO:0016818
GO:0003676
GO:0005515
GO:0016742
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
Nucleolus
Cytoplasm
Cytoskeleton
Spindle pole
Midbody
Microtubule organizing center
Centrosome
Chromosome (Microbial infection) Colocalizes with bovine papillomavirus type 1 regulatory protein E2 on mitotic chromosomes at early stages of mitosis. With evidence from 7 publications.
Nucleolus
Cytoplasm
Cytoskeleton
Spindle pole
Midbody
Microtubule organizing center
Centrosome
Chromosome (Microbial infection) Colocalizes with bovine papillomavirus type 1 regulatory protein E2 on mitotic chromosomes at early stages of mitosis. With evidence from 7 publications.
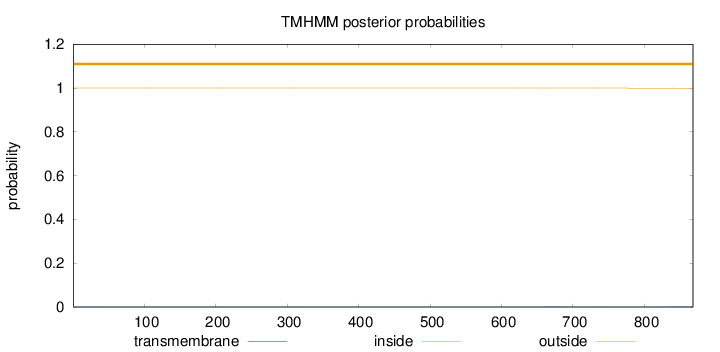
Length:
868
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02186
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.00006
outside
1 - 868
Population Genetic Test Statistics
Pi
217.94451
Theta
195.549284
Tajima's D
1.518176
CLR
12.265427
CSRT
0.7961601919904
Interpretation
Uncertain
