Pre Gene Modal
BGIBMGA008086
Annotation
PREDICTED:_dynactin_subunit_6_[Amyelois_transitella]
Full name
Polypeptide N-acetylgalactosaminyltransferase
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
Location in the cell
Cytoplasmic Reliability : 1.901
Sequence
CDS
ATGGCTCACAACATTAAAATTCTACCCGGAGCAACTGTCTGCGAAGACTGTACCTTAGAGGGTGATATAACCATAGGAGGAGGTACAGTTATTCATCCGAGAGTCAGCATTATAGCTGAAGGCGGACCTATTATAATTGCTGAATACTGTATTATTGAAGAATATTCTACTATAATACATAAAAAGAGTGATAAACAGGAGAATCCCCCAAAGCCATTGTTTATAGGTGCACACAATGTTTTTGAAGTTGCTTGCAAACTGGAGTCTATCTATGGTCACGTCGGTGAAAGTAATGTTTTTGAATGCAGATCTTTTGTTGGAGAAGAAGTCAAAGTTGGCAGTGGTTGTGTAATAGGAGCAGCCTGTACATTAACAGCTCCCCAGATATTAGCAGACAATACAGTGATCTGGGGCTCAGAGCATCATGTTAGAGAAGCTTTAGAGAAACAGCCCTCTCAGTTATTGCAGCTTGATTTCTTAAGCAAAGTGATGCCTAATTATCATAGACTGCGAAAACCAAATGTTCACAAAAGACAACCATCAAGACAGTCTCAAGAACCTTCTCCAAAACCTTAG
Protein
MAHNIKILPGATVCEDCTLEGDITIGGGTVIHPRVSIIAEGGPIIIAEYCIIEEYSTIIHKKSDKQENPPKPLFIGAHNVFEVACKLESIYGHVGESNVFECRSFVGEEVKVGSGCVIGAACTLTAPQILADNTVIWGSEHHVREALEKQPSQLLQLDFLSKVMPNYHRLRKPNVHKRQPSRQSQEPSPKP
Summary
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Feature
chain Polypeptide N-acetylgalactosaminyltransferase
Uniprot
H9JEY9
A0A2H1W361
A0A1E1WRK3
A0A2W1BS26
A0A2A4J8S3
S4NSV1
+ More
A0A212FLK6 A0A0N1I9X8 A0A194QM31 A0A0L7KKI8 B4KKF9 A0A1I8NPA2 B4JPA5 B4M940 A0A1W4XN15 A0A067R469 A0A1B0CGT8 A0A182QIC0 A0A023EID8 A0A182I6P5 A0A182V2Y9 A0A1J1HJJ8 A7URW0 A0A084WKP6 A0A182L3P4 D6WRV9 A0A1L8DVM2 A0A1L8DVN9 A0A1I8MKE8 A0A182K6W7 A0A182N9C4 A0A182IKH2 A0A336MWP2 A0A3B0KBI5 A0A0B6Y2S9 A0A1Y1KZ89 A0A182WWG5 A0A182Y293 A0A182RJJ3 A0A182WBG6 A0A182LYS9 A0A182PIK7 A0A182TH61 A0A2M4AY81 A0A1A9V9J3 A0A1B0D0N3 N6TJZ9 B0WWZ7 A0A1Q3FGF8 B0XFD0 A0A182HB50 A0A1A9W061 A0A2G8KY15 A0A1W4VXM1 B4MTI9 A0A1B0B7M8 A0A0A1X463 A0A2C9JJI6 Q16F67 B3NM85 Q6XIZ9 A0A2M4BYW0 A0A0R1DPD6 A0A0Q5VWB3 W5JIV6 B4GXE7 D0QWD1 U5EX38 B3MNN7 A0A034WN87 A0A0C9PU78 A0A0K8WI29 A0A1L8EDE2 Q29CK5 A0A1Y1KY99 A0A0A9WBJ5 B4I5S2 A0A1L8EDD9 B4Q9H3 V3ZEY3 Q9VIZ1 A0A2R7WVI7 A0A154P9I7 A0A2P2HZE9 A0A0J7KP48 A0A267GVE2 A0A267GSG7 A0A267F484 A0A151X660 A0A026VV08 A0A0M5JAU4 A0A195F4D2 A0A195B9E3 A0A182FEH6 A0A0B7BTJ2 A0A2L2YEP1 A0A2L2YEB5 A0A091E1G1 A0A0V0G883 A0A1B6E4V0
A0A212FLK6 A0A0N1I9X8 A0A194QM31 A0A0L7KKI8 B4KKF9 A0A1I8NPA2 B4JPA5 B4M940 A0A1W4XN15 A0A067R469 A0A1B0CGT8 A0A182QIC0 A0A023EID8 A0A182I6P5 A0A182V2Y9 A0A1J1HJJ8 A7URW0 A0A084WKP6 A0A182L3P4 D6WRV9 A0A1L8DVM2 A0A1L8DVN9 A0A1I8MKE8 A0A182K6W7 A0A182N9C4 A0A182IKH2 A0A336MWP2 A0A3B0KBI5 A0A0B6Y2S9 A0A1Y1KZ89 A0A182WWG5 A0A182Y293 A0A182RJJ3 A0A182WBG6 A0A182LYS9 A0A182PIK7 A0A182TH61 A0A2M4AY81 A0A1A9V9J3 A0A1B0D0N3 N6TJZ9 B0WWZ7 A0A1Q3FGF8 B0XFD0 A0A182HB50 A0A1A9W061 A0A2G8KY15 A0A1W4VXM1 B4MTI9 A0A1B0B7M8 A0A0A1X463 A0A2C9JJI6 Q16F67 B3NM85 Q6XIZ9 A0A2M4BYW0 A0A0R1DPD6 A0A0Q5VWB3 W5JIV6 B4GXE7 D0QWD1 U5EX38 B3MNN7 A0A034WN87 A0A0C9PU78 A0A0K8WI29 A0A1L8EDE2 Q29CK5 A0A1Y1KY99 A0A0A9WBJ5 B4I5S2 A0A1L8EDD9 B4Q9H3 V3ZEY3 Q9VIZ1 A0A2R7WVI7 A0A154P9I7 A0A2P2HZE9 A0A0J7KP48 A0A267GVE2 A0A267GSG7 A0A267F484 A0A151X660 A0A026VV08 A0A0M5JAU4 A0A195F4D2 A0A195B9E3 A0A182FEH6 A0A0B7BTJ2 A0A2L2YEP1 A0A2L2YEB5 A0A091E1G1 A0A0V0G883 A0A1B6E4V0
EC Number
2.4.1.-
Pubmed
19121390
28756777
23622113
22118469
26354079
26227816
+ More
17994087 24845553 24945155 12364791 24438588 20966253 18362917 19820115 25315136 28004739 25244985 23537049 26483478 29023486 25830018 15562597 17510324 14525923 17550304 20920257 23761445 19859648 25348373 15632085 25401762 26823975 22936249 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 26561354
17994087 24845553 24945155 12364791 24438588 20966253 18362917 19820115 25315136 28004739 25244985 23537049 26483478 29023486 25830018 15562597 17510324 14525923 17550304 20920257 23761445 19859648 25348373 15632085 25401762 26823975 22936249 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 26561354
EMBL
BABH01023645
ODYU01006040
SOQ47535.1
GDQN01001623
JAT89431.1
KZ149974
+ More
PZC75977.1 NWSH01002634 PCG67793.1 GAIX01012421 JAA80139.1 AGBW02007748 OWR54616.1 KQ459330 KQ459144 KPJ01435.1 KPJ03704.1 KQ461198 KPJ06015.1 JTDY01009382 KOB63586.1 CH933807 EDW11609.1 CH916372 EDV98735.1 CH940654 EDW57716.1 KK852711 KDR17944.1 AJWK01011585 AXCN02000559 GAPW01004731 JAC08867.1 APCN01003688 CVRI01000006 CRK88232.1 AAAB01008807 EDO64752.1 ATLV01024132 KE525349 KFB50790.1 KQ971351 EFA07650.2 GFDF01003593 JAV10491.1 GFDF01003594 JAV10490.1 UFQT01001397 SSX30368.1 OUUW01000010 SPP85520.1 HACG01003554 CEK50419.1 GEZM01074119 GEZM01074117 JAV64666.1 AXCM01008947 GGFK01012426 MBW45747.1 AJVK01010006 APGK01035251 KB740923 KB632333 ENN78223.1 ERL92658.1 DS232155 EDS36258.1 GFDL01008409 JAV26636.1 DS232914 EDS26703.1 JXUM01031699 KQ560932 KXJ80297.1 MRZV01000310 PIK52913.1 CH963852 EDW75428.1 JXJN01009696 GBXI01008415 JAD05877.1 CH478504 CH478064 EAT32878.1 EAT34138.1 CH954179 EDV54685.1 AY231680 CM000158 AAR09703.1 EDW90385.1 GGFJ01009108 MBW58249.1 KRJ99146.1 KQS61902.1 ADMH02001370 ETN62820.1 CH479195 EDW27258.1 FJ821025 ACN94630.1 GANO01002696 JAB57175.1 CH902620 EDV31124.1 GAKP01003751 GAKP01003749 JAC55203.1 GBYB01004903 JAG74670.1 GDHF01001774 JAI50540.1 GFDG01002048 JAV16751.1 CH475456 EAL29382.3 GEZM01074118 JAV64665.1 GBHO01038490 GBRD01012099 GDHC01012980 JAG05114.1 JAG53725.1 JAQ05649.1 CH480822 EDW55728.1 GFDG01002049 JAV16750.1 CM000361 CM002910 EDX05480.1 KMY90953.1 KB202544 ESO89713.1 AE014134 BT023827 AAF53769.1 AAZ86748.1 KK855693 PTY23558.1 KQ434849 KZC08501.1 IACF01001432 LAB67133.1 LBMM01004775 KMQ92097.1 NIVC01000153 PAA89317.1 NIVC01000169 PAA88966.1 NIVC01001383 PAA68590.1 KQ982482 KYQ55871.1 KK107842 QOIP01000003 EZA47515.1 RLU24838.1 CP012525 ALC42868.1 KQ981836 KYN35032.1 KQ976542 KYM81158.1 HACG01049649 CEK96514.1 IAAA01024757 LAA06487.1 IAAA01024758 LAA06491.1 KN120606 KFO38184.1 GECL01001899 JAP04225.1 GEDC01004336 JAS32962.1
PZC75977.1 NWSH01002634 PCG67793.1 GAIX01012421 JAA80139.1 AGBW02007748 OWR54616.1 KQ459330 KQ459144 KPJ01435.1 KPJ03704.1 KQ461198 KPJ06015.1 JTDY01009382 KOB63586.1 CH933807 EDW11609.1 CH916372 EDV98735.1 CH940654 EDW57716.1 KK852711 KDR17944.1 AJWK01011585 AXCN02000559 GAPW01004731 JAC08867.1 APCN01003688 CVRI01000006 CRK88232.1 AAAB01008807 EDO64752.1 ATLV01024132 KE525349 KFB50790.1 KQ971351 EFA07650.2 GFDF01003593 JAV10491.1 GFDF01003594 JAV10490.1 UFQT01001397 SSX30368.1 OUUW01000010 SPP85520.1 HACG01003554 CEK50419.1 GEZM01074119 GEZM01074117 JAV64666.1 AXCM01008947 GGFK01012426 MBW45747.1 AJVK01010006 APGK01035251 KB740923 KB632333 ENN78223.1 ERL92658.1 DS232155 EDS36258.1 GFDL01008409 JAV26636.1 DS232914 EDS26703.1 JXUM01031699 KQ560932 KXJ80297.1 MRZV01000310 PIK52913.1 CH963852 EDW75428.1 JXJN01009696 GBXI01008415 JAD05877.1 CH478504 CH478064 EAT32878.1 EAT34138.1 CH954179 EDV54685.1 AY231680 CM000158 AAR09703.1 EDW90385.1 GGFJ01009108 MBW58249.1 KRJ99146.1 KQS61902.1 ADMH02001370 ETN62820.1 CH479195 EDW27258.1 FJ821025 ACN94630.1 GANO01002696 JAB57175.1 CH902620 EDV31124.1 GAKP01003751 GAKP01003749 JAC55203.1 GBYB01004903 JAG74670.1 GDHF01001774 JAI50540.1 GFDG01002048 JAV16751.1 CH475456 EAL29382.3 GEZM01074118 JAV64665.1 GBHO01038490 GBRD01012099 GDHC01012980 JAG05114.1 JAG53725.1 JAQ05649.1 CH480822 EDW55728.1 GFDG01002049 JAV16750.1 CM000361 CM002910 EDX05480.1 KMY90953.1 KB202544 ESO89713.1 AE014134 BT023827 AAF53769.1 AAZ86748.1 KK855693 PTY23558.1 KQ434849 KZC08501.1 IACF01001432 LAB67133.1 LBMM01004775 KMQ92097.1 NIVC01000153 PAA89317.1 NIVC01000169 PAA88966.1 NIVC01001383 PAA68590.1 KQ982482 KYQ55871.1 KK107842 QOIP01000003 EZA47515.1 RLU24838.1 CP012525 ALC42868.1 KQ981836 KYN35032.1 KQ976542 KYM81158.1 HACG01049649 CEK96514.1 IAAA01024757 LAA06487.1 IAAA01024758 LAA06491.1 KN120606 KFO38184.1 GECL01001899 JAP04225.1 GEDC01004336 JAS32962.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000009192 UP000095300 UP000001070 UP000008792 UP000192223 UP000027135 UP000092461 UP000075886 UP000075840 UP000075903 UP000183832 UP000007062 UP000030765 UP000075882 UP000007266 UP000095301 UP000075881 UP000075884 UP000075880 UP000268350 UP000076407 UP000076408 UP000075900 UP000075920 UP000075883 UP000075885 UP000075902 UP000078200 UP000092462 UP000019118 UP000030742 UP000002320 UP000069940 UP000249989 UP000091820 UP000230750 UP000192221 UP000007798 UP000092460 UP000076420 UP000008820 UP000008711 UP000002282 UP000000673 UP000008744 UP000007801 UP000001819 UP000001292 UP000000304 UP000030746 UP000000803 UP000076502 UP000036403 UP000215902 UP000075809 UP000053097 UP000279307 UP000092553 UP000078541 UP000078540 UP000069272 UP000028990
UP000009192 UP000095300 UP000001070 UP000008792 UP000192223 UP000027135 UP000092461 UP000075886 UP000075840 UP000075903 UP000183832 UP000007062 UP000030765 UP000075882 UP000007266 UP000095301 UP000075881 UP000075884 UP000075880 UP000268350 UP000076407 UP000076408 UP000075900 UP000075920 UP000075883 UP000075885 UP000075902 UP000078200 UP000092462 UP000019118 UP000030742 UP000002320 UP000069940 UP000249989 UP000091820 UP000230750 UP000192221 UP000007798 UP000092460 UP000076420 UP000008820 UP000008711 UP000002282 UP000000673 UP000008744 UP000007801 UP000001819 UP000001292 UP000000304 UP000030746 UP000000803 UP000076502 UP000036403 UP000215902 UP000075809 UP000053097 UP000279307 UP000092553 UP000078541 UP000078540 UP000069272 UP000028990
PRIDE
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEY9
A0A2H1W361
A0A1E1WRK3
A0A2W1BS26
A0A2A4J8S3
S4NSV1
+ More
A0A212FLK6 A0A0N1I9X8 A0A194QM31 A0A0L7KKI8 B4KKF9 A0A1I8NPA2 B4JPA5 B4M940 A0A1W4XN15 A0A067R469 A0A1B0CGT8 A0A182QIC0 A0A023EID8 A0A182I6P5 A0A182V2Y9 A0A1J1HJJ8 A7URW0 A0A084WKP6 A0A182L3P4 D6WRV9 A0A1L8DVM2 A0A1L8DVN9 A0A1I8MKE8 A0A182K6W7 A0A182N9C4 A0A182IKH2 A0A336MWP2 A0A3B0KBI5 A0A0B6Y2S9 A0A1Y1KZ89 A0A182WWG5 A0A182Y293 A0A182RJJ3 A0A182WBG6 A0A182LYS9 A0A182PIK7 A0A182TH61 A0A2M4AY81 A0A1A9V9J3 A0A1B0D0N3 N6TJZ9 B0WWZ7 A0A1Q3FGF8 B0XFD0 A0A182HB50 A0A1A9W061 A0A2G8KY15 A0A1W4VXM1 B4MTI9 A0A1B0B7M8 A0A0A1X463 A0A2C9JJI6 Q16F67 B3NM85 Q6XIZ9 A0A2M4BYW0 A0A0R1DPD6 A0A0Q5VWB3 W5JIV6 B4GXE7 D0QWD1 U5EX38 B3MNN7 A0A034WN87 A0A0C9PU78 A0A0K8WI29 A0A1L8EDE2 Q29CK5 A0A1Y1KY99 A0A0A9WBJ5 B4I5S2 A0A1L8EDD9 B4Q9H3 V3ZEY3 Q9VIZ1 A0A2R7WVI7 A0A154P9I7 A0A2P2HZE9 A0A0J7KP48 A0A267GVE2 A0A267GSG7 A0A267F484 A0A151X660 A0A026VV08 A0A0M5JAU4 A0A195F4D2 A0A195B9E3 A0A182FEH6 A0A0B7BTJ2 A0A2L2YEP1 A0A2L2YEB5 A0A091E1G1 A0A0V0G883 A0A1B6E4V0
A0A212FLK6 A0A0N1I9X8 A0A194QM31 A0A0L7KKI8 B4KKF9 A0A1I8NPA2 B4JPA5 B4M940 A0A1W4XN15 A0A067R469 A0A1B0CGT8 A0A182QIC0 A0A023EID8 A0A182I6P5 A0A182V2Y9 A0A1J1HJJ8 A7URW0 A0A084WKP6 A0A182L3P4 D6WRV9 A0A1L8DVM2 A0A1L8DVN9 A0A1I8MKE8 A0A182K6W7 A0A182N9C4 A0A182IKH2 A0A336MWP2 A0A3B0KBI5 A0A0B6Y2S9 A0A1Y1KZ89 A0A182WWG5 A0A182Y293 A0A182RJJ3 A0A182WBG6 A0A182LYS9 A0A182PIK7 A0A182TH61 A0A2M4AY81 A0A1A9V9J3 A0A1B0D0N3 N6TJZ9 B0WWZ7 A0A1Q3FGF8 B0XFD0 A0A182HB50 A0A1A9W061 A0A2G8KY15 A0A1W4VXM1 B4MTI9 A0A1B0B7M8 A0A0A1X463 A0A2C9JJI6 Q16F67 B3NM85 Q6XIZ9 A0A2M4BYW0 A0A0R1DPD6 A0A0Q5VWB3 W5JIV6 B4GXE7 D0QWD1 U5EX38 B3MNN7 A0A034WN87 A0A0C9PU78 A0A0K8WI29 A0A1L8EDE2 Q29CK5 A0A1Y1KY99 A0A0A9WBJ5 B4I5S2 A0A1L8EDD9 B4Q9H3 V3ZEY3 Q9VIZ1 A0A2R7WVI7 A0A154P9I7 A0A2P2HZE9 A0A0J7KP48 A0A267GVE2 A0A267GSG7 A0A267F484 A0A151X660 A0A026VV08 A0A0M5JAU4 A0A195F4D2 A0A195B9E3 A0A182FEH6 A0A0B7BTJ2 A0A2L2YEP1 A0A2L2YEB5 A0A091E1G1 A0A0V0G883 A0A1B6E4V0
PDB
6F3A
E-value=6.28595e-29,
Score=313
Ontologies
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
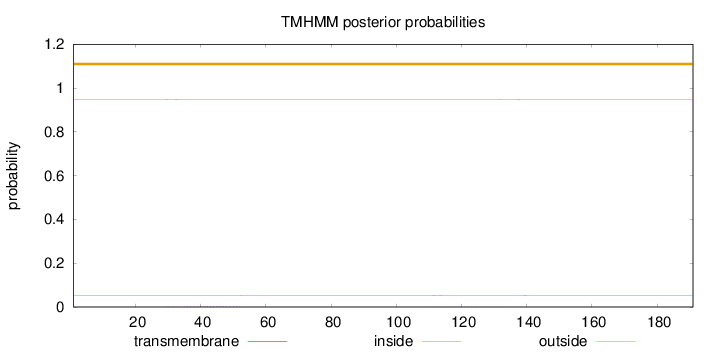
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0731500000000001
Exp number, first 60 AAs:
0.05441
Total prob of N-in:
0.05145
outside
1 - 191
Population Genetic Test Statistics
Pi
197.499266
Theta
181.758262
Tajima's D
0.100223
CLR
3.530882
CSRT
0.395980200989951
Interpretation
Uncertain
