Gene
KWMTBOMO04969 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007986
Annotation
PREDICTED:_phosphatidylinositol_4-kinase_beta_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.115
Sequence
CDS
ATGTTGGAGGATGTTCCCGTGCACGGAAGTCGGTCAGACTCCCGCAATGGAAAGAGTACTATCCACCAGAGAAACCTCAGCTTGGATTTCAGGCATGTGACAGGCGCAGGAGGGCCGCCAGCGGGATGCGGTCATCAAAGGAACAGGTCCCTGGACTCTGTGCTCCAACGGATACCAGAAGACATGACACCAACTTCGCCGGAGGATGCACCTCTGCGACTCACTATCGAAGCGACGGTACCAAGATTGGCGCTACCCCCGGAAGTGCCGACAGGCTCAGACGACTCTGGTATTCACACGCCGCCCGATGACAACGAAGACGAGCGACCCCCGCTCCCGACAGCGGCGCGGTCTAGAGAAAGTCTTGATTCTGGAAATCCGGAAGACACAGAGAAACCTCCCGATCCCCCCGATACGATAGCAGAACCGACAACTACAGCTGGCACCAGCGTCGCAAGCACGTCCAAAAACAAAGACTCTCTGTTAAGACTATTCGAGAGTTCGTTGTTTGACATGAACATGGCAATAGAATATTTTCACAACTCCTCGGGCGTACAGACTTATCTGGCGAATAAGTTGTTTTCGTTTCCGCACGAGGATGTCGATTTTTATCTGCCGCAGCTGGCAACGATGTACATAGAGTTAGCCGACGTAGCGGAGGTGCTAAAGCCGTATTTGATTCATAGATGTAAACAGAGCACGGAGTTTTCGCTGCGACTCGTATGGTTGTTGACCGCGTTCGGTGGAGATTCGACAACGTCTAAAGCGATGAAGCTGAGAGAATCCATACTGGCCGAAAAGATTAGGCCGGCAGCGAAAAGAGGACATCACCGAACACAATCGGACGCTTCGGCGTTGAGAGTGTCGACGCCTCCTTGGAAATATTTAGGGGACTTGACATCTGGACGGGCCTTCGATAATGGTTGTGTTTGTGGGGAACAGTGTTCGTGCGACGCTCCCAGACTGGCACCGCATTTAGAGTTCATGTTGGCGCTAATGAATATCGGACGCCGGTTGGCCAGCGTCGCGGACAAGGAACGAAGGAATGGCCACCTCAGAGCTGAACTAGCGACTCTGAACCTGAATTTGCCGGCCAAAGTTTGGCTACCCCTATATCACAAGCCTCACTACGTGTTGAGGATATCACCGTACGCGGCCACGGTGCTCAATAGCAAAGATAAGGCTCCGTACATAATGTACGTGGAGGTACTAGAGACTGACGGGCACGAGCCTCTTCCTCCTCGCCAGCTGCCTCCAGCGCCACCAGCTCCCCACTCTCCCCCCATCAGACATACGAAGAGTGAGGAGAACCTCGTGCCTGAGTGGCCTGCGCTCTCCCTGTATTCGGCACTGGACGACGTGGACACCGGAGACTGCTGGAGTCATGACGACGATGAACTCAGCTCTCATTACGACCACAGAGCGCCCGCGCCCGACAGCGTGTCGCAGGTCTCGGCGGTTTCGGTTCTGTCCACGTCATCGTGCGGCAGCCGCGAGTCTGTGGCCGTGGTCGCCGGCGAGGTGCGCAGGAGGCTGGTCGACGCCACAGAAGCAGAAACCTTCAGACACGACCCCGCCGATCCGTCCGCTACCGCACTTAAGGAATCGTGGGAGAGCAAGCTGTGCCGCATCCGCGCCATGTCCCCGTACGGCGCGCTGTCGGGCTGGCGGCTGCTGGGCTGCATCGTGAAGGTGGGCGACGACCTGCGCCAGGAGCTGCTGGCCGCGCAGCTGCTGCGCCGCCTCACCGCCATCTGGCGCCACGAGCGCGTGCCGCTGGCGCTGCGCCCCTACGAGATCCTGTGTTTGGGCCGCGACACCGGACTCATACAACCGGTGCTGAACTCTGTGTCGCTGCACCAGATCAAGAAGCAGTCGGGGAAGTCCCTGCGCGCGTGGCTACAGCTAGAGCACGGCGCGCCCACCTCTGAAGGCTTCCTGCGCGCGCAAAGGAACTTCGTGGAATCCGTCGCCGCCTACGCGCTCGCCAGCTATCTGCTGCAACTCAAGGACCGGCACAACGGAAACATCTTATTGGACAGCGAAGGTCACATAATCCACATCGACTTCGGGTTCATATTCTCTACGTCACCGAAAAATCTTGGATTCGAAAGTTCACCGTTCAAATTGACACAGGAGTTCGTTGAGGTAATGGACGGCGAACACTCGGACATGTTTCAATACTTCAAGATATTGATCCTGCGCGGCCTCATCGCGGCGCGCAAGTACTGCGACCAGATCATACACATGGTCGAGCTGATGCGAATGAGCGGTCAGCTGGCGTGCCTCCGCAGTAGTAGCGTGGTGTCGTCGTTCCGCGCTCGCTTCCAGAGCGGCAAGACCGACCCGCAGCTGCAGGCGCTCGTCGACAGGCTCGTGCAGGACGCGCTCAACTCCATCTCCACCAGGCTCTACGACAACTACCAGTACCACACCAACGGGATACTGTGA
Protein
MLEDVPVHGSRSDSRNGKSTIHQRNLSLDFRHVTGAGGPPAGCGHQRNRSLDSVLQRIPEDMTPTSPEDAPLRLTIEATVPRLALPPEVPTGSDDSGIHTPPDDNEDERPPLPTAARSRESLDSGNPEDTEKPPDPPDTIAEPTTTAGTSVASTSKNKDSLLRLFESSLFDMNMAIEYFHNSSGVQTYLANKLFSFPHEDVDFYLPQLATMYIELADVAEVLKPYLIHRCKQSTEFSLRLVWLLTAFGGDSTTSKAMKLRESILAEKIRPAAKRGHHRTQSDASALRVSTPPWKYLGDLTSGRAFDNGCVCGEQCSCDAPRLAPHLEFMLALMNIGRRLASVADKERRNGHLRAELATLNLNLPAKVWLPLYHKPHYVLRISPYAATVLNSKDKAPYIMYVEVLETDGHEPLPPRQLPPAPPAPHSPPIRHTKSEENLVPEWPALSLYSALDDVDTGDCWSHDDDELSSHYDHRAPAPDSVSQVSAVSVLSTSSCGSRESVAVVAGEVRRRLVDATEAETFRHDPADPSATALKESWESKLCRIRAMSPYGALSGWRLLGCIVKVGDDLRQELLAAQLLRRLTAIWRHERVPLALRPYEILCLGRDTGLIQPVLNSVSLHQIKKQSGKSLRAWLQLEHGAPTSEGFLRAQRNFVESVAAYALASYLLQLKDRHNGNILLDSEGHIIHIDFGFIFSTSPKNLGFESSPFKLTQEFVEVMDGEHSDMFQYFKILILRGLIAARKYCDQIIHMVELMRMSGQLACLRSSSVVSSFRARFQSGKTDPQLQALVDRLVQDALNSISTRLYDNYQYHTNGIL
Summary
Similarity
Belongs to the PI3/PI4-kinase family.
Uniprot
A0A2W1BTL8
A0A2H1VT94
A0A194QKU1
A0A194QDS1
A0A212FLL2
A0A146KRY8
+ More
A0A0K8SAJ8 D6X462 A0A0A9YXT1 A0A023F2H6 A0A224XJA5 A0A069DX32 A0A2P8Y5H7 A0A067QLB2 A0A2R7W4C9 T1IHS6 A0A0P5RML4 A0A0P5QGF9 A0A0P5MXL3 A0A0P5K416 A0A0P5SJF3 A0A164LGB7 A0A0P5CQE1 A0A0P5EBW3 A0A0P4Z5M5 A0A0N8C4E2 A0A1Y1LSC9 A0A0P6BUX1 A0A0P4ZYG3 A0A0N8CK46 A0A0P5Y2G1 A0A0P5EAD9 A0A1Y1LQE4 A0A0P5UL37 A0A0N8EPZ9 A0A0N8C9S8 A0A0P4Z7S4 E9J1K6 A0A195CP02 A0A0J7L901 E2AM46 A0A1S3HHC1 A0A1W4WH15 E9H0X8 A0A1E1X2D8 A0A147BS57 A0A131XWG7 A0A3S3PF60 V5HKJ4 A0A131YKX6 A0A224Z436 L7M037 A0A2T7PFL5 A0A0P5WB41 A0A1Z5L4N8 A0A131XIV9 A0A1D2MN43 A0A2R5LNV9 A0A210QMT2 A0A2L2Y9S4 A0A2P6LCB9 A0A0L8GQJ1 A0A3Q1J332 A0A0P5M7I2 A0A087TWF8 A0A3Q0RV42 W4YK59 A0A226DG11 A0A3B5A115 A0A3B4WBX5 A0A3Q1IZ07 A0A0F8ANZ4 A0A3B4UMU1 A0A147ALK0 A0A3P8T0Z7 A0A3Q2V9F6 A0A3P8Q245 V3ZYP0 A0A3B5PRL2 A0A3Q4GV66 A0A3B3BD70 A0A3Q1BXK5 A0A3B3BCB4 A0A3B4GCA7 A0A3Q2WJZ3 A0A3Q3SJ43 A0A3B4EH15 A0A3B5AL65 A0A3P9HZK1 A0A3Q2FTK0 H2N2X9 A0A3B3TUW3 H2SZZ4 A0A3Q3JR25 A0A3Q1G5A1 A0A146XPU0 A0A3B3SIX8 A0A1A7X800
A0A0K8SAJ8 D6X462 A0A0A9YXT1 A0A023F2H6 A0A224XJA5 A0A069DX32 A0A2P8Y5H7 A0A067QLB2 A0A2R7W4C9 T1IHS6 A0A0P5RML4 A0A0P5QGF9 A0A0P5MXL3 A0A0P5K416 A0A0P5SJF3 A0A164LGB7 A0A0P5CQE1 A0A0P5EBW3 A0A0P4Z5M5 A0A0N8C4E2 A0A1Y1LSC9 A0A0P6BUX1 A0A0P4ZYG3 A0A0N8CK46 A0A0P5Y2G1 A0A0P5EAD9 A0A1Y1LQE4 A0A0P5UL37 A0A0N8EPZ9 A0A0N8C9S8 A0A0P4Z7S4 E9J1K6 A0A195CP02 A0A0J7L901 E2AM46 A0A1S3HHC1 A0A1W4WH15 E9H0X8 A0A1E1X2D8 A0A147BS57 A0A131XWG7 A0A3S3PF60 V5HKJ4 A0A131YKX6 A0A224Z436 L7M037 A0A2T7PFL5 A0A0P5WB41 A0A1Z5L4N8 A0A131XIV9 A0A1D2MN43 A0A2R5LNV9 A0A210QMT2 A0A2L2Y9S4 A0A2P6LCB9 A0A0L8GQJ1 A0A3Q1J332 A0A0P5M7I2 A0A087TWF8 A0A3Q0RV42 W4YK59 A0A226DG11 A0A3B5A115 A0A3B4WBX5 A0A3Q1IZ07 A0A0F8ANZ4 A0A3B4UMU1 A0A147ALK0 A0A3P8T0Z7 A0A3Q2V9F6 A0A3P8Q245 V3ZYP0 A0A3B5PRL2 A0A3Q4GV66 A0A3B3BD70 A0A3Q1BXK5 A0A3B3BCB4 A0A3B4GCA7 A0A3Q2WJZ3 A0A3Q3SJ43 A0A3B4EH15 A0A3B5AL65 A0A3P9HZK1 A0A3Q2FTK0 H2N2X9 A0A3B3TUW3 H2SZZ4 A0A3Q3JR25 A0A3Q1G5A1 A0A146XPU0 A0A3B3SIX8 A0A1A7X800
Pubmed
28756777
26354079
22118469
26823975
18362917
19820115
+ More
25401762 25474469 26334808 29403074 24845553 28004739 21282665 20798317 21292972 28503490 29652888 25765539 26830274 28797301 25576852 28528879 28049606 27289101 28812685 26561354 25835551 23254933 23542700 25186727 29451363 17554307 21551351 29240929
25401762 25474469 26334808 29403074 24845553 28004739 21282665 20798317 21292972 28503490 29652888 25765539 26830274 28797301 25576852 28528879 28049606 27289101 28812685 26561354 25835551 23254933 23542700 25186727 29451363 17554307 21551351 29240929
EMBL
KZ149974
PZC75976.1
ODYU01004324
SOQ44071.1
KQ461198
KPJ06014.1
+ More
KQ459144 KPJ03703.1 AGBW02007748 OWR54617.1 GDHC01019365 GDHC01001507 JAP99263.1 JAQ17122.1 GBRD01015519 JAG50307.1 KQ971379 EEZ97712.1 GBHO01009229 GBHO01009228 JAG34375.1 JAG34376.1 GBBI01003207 JAC15505.1 GFTR01008213 JAW08213.1 GBGD01000627 JAC88262.1 PYGN01000904 PSN39507.1 KK853204 KDR09872.1 KK854317 PTY14584.1 JH430009 GDIQ01117474 JAL34252.1 GDIQ01117473 JAL34253.1 GDIQ01150354 JAL01372.1 GDIQ01215586 JAK36139.1 GDIQ01092430 JAL59296.1 LRGB01003134 KZS04083.1 GDIP01170966 JAJ52436.1 GDIP01150126 JAJ73276.1 GDIP01218269 JAJ05133.1 GDIQ01115923 JAL35803.1 GEZM01049748 JAV75881.1 GDIP01009821 JAM93894.1 GDIP01219562 JAJ03840.1 GDIP01123879 JAL79835.1 GDIP01063896 JAM39819.1 GDIP01145923 JAJ77479.1 GEZM01049747 JAV75882.1 GDIP01111380 JAL92334.1 GDIQ01005276 JAN89461.1 GDIQ01100835 JAL50891.1 GDIP01218270 JAJ05132.1 GL767674 EFZ13183.1 KQ977513 KYN02212.1 LBMM01000206 KMR04490.1 GL440704 EFN65504.1 GL732582 EFX74536.1 GFAC01005761 JAT93427.1 GEGO01001798 JAR93606.1 GEFM01005875 JAP69921.1 NCKU01005431 NCKU01001796 RWS04579.1 RWS11228.1 GANP01010490 JAB73978.1 GEDV01009831 JAP78726.1 GFPF01010545 MAA21691.1 GACK01007358 JAA57676.1 PZQS01000004 PVD32190.1 GDIP01089729 JAM13986.1 GFJQ02004597 JAW02373.1 GEFH01002124 JAP66457.1 LJIJ01000818 ODM94358.1 GGLE01006982 MBY11108.1 NEDP02002776 OWF50033.1 IAAA01011331 IAAA01011332 LAA04901.1 MWRG01000386 PRD36223.1 KQ420917 KOF78880.1 GDIQ01171546 JAK80179.1 KK117072 KFM69447.1 AAGJ04105879 AAGJ04105880 AAGJ04105881 AAGJ04105882 AAGJ04105883 LNIX01000019 OXA44482.1 KQ041575 KKF24722.1 GCES01006917 JAR79406.1 KB203188 ESO86106.1 GCES01042220 JAR44103.1 HADW01012539 SBP13939.1
KQ459144 KPJ03703.1 AGBW02007748 OWR54617.1 GDHC01019365 GDHC01001507 JAP99263.1 JAQ17122.1 GBRD01015519 JAG50307.1 KQ971379 EEZ97712.1 GBHO01009229 GBHO01009228 JAG34375.1 JAG34376.1 GBBI01003207 JAC15505.1 GFTR01008213 JAW08213.1 GBGD01000627 JAC88262.1 PYGN01000904 PSN39507.1 KK853204 KDR09872.1 KK854317 PTY14584.1 JH430009 GDIQ01117474 JAL34252.1 GDIQ01117473 JAL34253.1 GDIQ01150354 JAL01372.1 GDIQ01215586 JAK36139.1 GDIQ01092430 JAL59296.1 LRGB01003134 KZS04083.1 GDIP01170966 JAJ52436.1 GDIP01150126 JAJ73276.1 GDIP01218269 JAJ05133.1 GDIQ01115923 JAL35803.1 GEZM01049748 JAV75881.1 GDIP01009821 JAM93894.1 GDIP01219562 JAJ03840.1 GDIP01123879 JAL79835.1 GDIP01063896 JAM39819.1 GDIP01145923 JAJ77479.1 GEZM01049747 JAV75882.1 GDIP01111380 JAL92334.1 GDIQ01005276 JAN89461.1 GDIQ01100835 JAL50891.1 GDIP01218270 JAJ05132.1 GL767674 EFZ13183.1 KQ977513 KYN02212.1 LBMM01000206 KMR04490.1 GL440704 EFN65504.1 GL732582 EFX74536.1 GFAC01005761 JAT93427.1 GEGO01001798 JAR93606.1 GEFM01005875 JAP69921.1 NCKU01005431 NCKU01001796 RWS04579.1 RWS11228.1 GANP01010490 JAB73978.1 GEDV01009831 JAP78726.1 GFPF01010545 MAA21691.1 GACK01007358 JAA57676.1 PZQS01000004 PVD32190.1 GDIP01089729 JAM13986.1 GFJQ02004597 JAW02373.1 GEFH01002124 JAP66457.1 LJIJ01000818 ODM94358.1 GGLE01006982 MBY11108.1 NEDP02002776 OWF50033.1 IAAA01011331 IAAA01011332 LAA04901.1 MWRG01000386 PRD36223.1 KQ420917 KOF78880.1 GDIQ01171546 JAK80179.1 KK117072 KFM69447.1 AAGJ04105879 AAGJ04105880 AAGJ04105881 AAGJ04105882 AAGJ04105883 LNIX01000019 OXA44482.1 KQ041575 KKF24722.1 GCES01006917 JAR79406.1 KB203188 ESO86106.1 GCES01042220 JAR44103.1 HADW01012539 SBP13939.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000007266
UP000245037
UP000027135
+ More
UP000076858 UP000078542 UP000036403 UP000000311 UP000085678 UP000192223 UP000000305 UP000285301 UP000245119 UP000094527 UP000242188 UP000053454 UP000265040 UP000054359 UP000261340 UP000007110 UP000198287 UP000261400 UP000261360 UP000261420 UP000265080 UP000264840 UP000265100 UP000030746 UP000002852 UP000261580 UP000261560 UP000257160 UP000261460 UP000261640 UP000261440 UP000265200 UP000265020 UP000001038 UP000261500 UP000005226 UP000261600 UP000257200 UP000261540
UP000076858 UP000078542 UP000036403 UP000000311 UP000085678 UP000192223 UP000000305 UP000285301 UP000245119 UP000094527 UP000242188 UP000053454 UP000265040 UP000054359 UP000261340 UP000007110 UP000198287 UP000261400 UP000261360 UP000261420 UP000265080 UP000264840 UP000265100 UP000030746 UP000002852 UP000261580 UP000261560 UP000257160 UP000261460 UP000261640 UP000261440 UP000265200 UP000265020 UP000001038 UP000261500 UP000005226 UP000261600 UP000257200 UP000261540
Pfam
PF00454 PI3_PI4_kinase
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BTL8
A0A2H1VT94
A0A194QKU1
A0A194QDS1
A0A212FLL2
A0A146KRY8
+ More
A0A0K8SAJ8 D6X462 A0A0A9YXT1 A0A023F2H6 A0A224XJA5 A0A069DX32 A0A2P8Y5H7 A0A067QLB2 A0A2R7W4C9 T1IHS6 A0A0P5RML4 A0A0P5QGF9 A0A0P5MXL3 A0A0P5K416 A0A0P5SJF3 A0A164LGB7 A0A0P5CQE1 A0A0P5EBW3 A0A0P4Z5M5 A0A0N8C4E2 A0A1Y1LSC9 A0A0P6BUX1 A0A0P4ZYG3 A0A0N8CK46 A0A0P5Y2G1 A0A0P5EAD9 A0A1Y1LQE4 A0A0P5UL37 A0A0N8EPZ9 A0A0N8C9S8 A0A0P4Z7S4 E9J1K6 A0A195CP02 A0A0J7L901 E2AM46 A0A1S3HHC1 A0A1W4WH15 E9H0X8 A0A1E1X2D8 A0A147BS57 A0A131XWG7 A0A3S3PF60 V5HKJ4 A0A131YKX6 A0A224Z436 L7M037 A0A2T7PFL5 A0A0P5WB41 A0A1Z5L4N8 A0A131XIV9 A0A1D2MN43 A0A2R5LNV9 A0A210QMT2 A0A2L2Y9S4 A0A2P6LCB9 A0A0L8GQJ1 A0A3Q1J332 A0A0P5M7I2 A0A087TWF8 A0A3Q0RV42 W4YK59 A0A226DG11 A0A3B5A115 A0A3B4WBX5 A0A3Q1IZ07 A0A0F8ANZ4 A0A3B4UMU1 A0A147ALK0 A0A3P8T0Z7 A0A3Q2V9F6 A0A3P8Q245 V3ZYP0 A0A3B5PRL2 A0A3Q4GV66 A0A3B3BD70 A0A3Q1BXK5 A0A3B3BCB4 A0A3B4GCA7 A0A3Q2WJZ3 A0A3Q3SJ43 A0A3B4EH15 A0A3B5AL65 A0A3P9HZK1 A0A3Q2FTK0 H2N2X9 A0A3B3TUW3 H2SZZ4 A0A3Q3JR25 A0A3Q1G5A1 A0A146XPU0 A0A3B3SIX8 A0A1A7X800
A0A0K8SAJ8 D6X462 A0A0A9YXT1 A0A023F2H6 A0A224XJA5 A0A069DX32 A0A2P8Y5H7 A0A067QLB2 A0A2R7W4C9 T1IHS6 A0A0P5RML4 A0A0P5QGF9 A0A0P5MXL3 A0A0P5K416 A0A0P5SJF3 A0A164LGB7 A0A0P5CQE1 A0A0P5EBW3 A0A0P4Z5M5 A0A0N8C4E2 A0A1Y1LSC9 A0A0P6BUX1 A0A0P4ZYG3 A0A0N8CK46 A0A0P5Y2G1 A0A0P5EAD9 A0A1Y1LQE4 A0A0P5UL37 A0A0N8EPZ9 A0A0N8C9S8 A0A0P4Z7S4 E9J1K6 A0A195CP02 A0A0J7L901 E2AM46 A0A1S3HHC1 A0A1W4WH15 E9H0X8 A0A1E1X2D8 A0A147BS57 A0A131XWG7 A0A3S3PF60 V5HKJ4 A0A131YKX6 A0A224Z436 L7M037 A0A2T7PFL5 A0A0P5WB41 A0A1Z5L4N8 A0A131XIV9 A0A1D2MN43 A0A2R5LNV9 A0A210QMT2 A0A2L2Y9S4 A0A2P6LCB9 A0A0L8GQJ1 A0A3Q1J332 A0A0P5M7I2 A0A087TWF8 A0A3Q0RV42 W4YK59 A0A226DG11 A0A3B5A115 A0A3B4WBX5 A0A3Q1IZ07 A0A0F8ANZ4 A0A3B4UMU1 A0A147ALK0 A0A3P8T0Z7 A0A3Q2V9F6 A0A3P8Q245 V3ZYP0 A0A3B5PRL2 A0A3Q4GV66 A0A3B3BD70 A0A3Q1BXK5 A0A3B3BCB4 A0A3B4GCA7 A0A3Q2WJZ3 A0A3Q3SJ43 A0A3B4EH15 A0A3B5AL65 A0A3P9HZK1 A0A3Q2FTK0 H2N2X9 A0A3B3TUW3 H2SZZ4 A0A3Q3JR25 A0A3Q1G5A1 A0A146XPU0 A0A3B3SIX8 A0A1A7X800
PDB
5EUQ
E-value=3.14272e-71,
Score=685
Ontologies
PATHWAY
GO
PANTHER
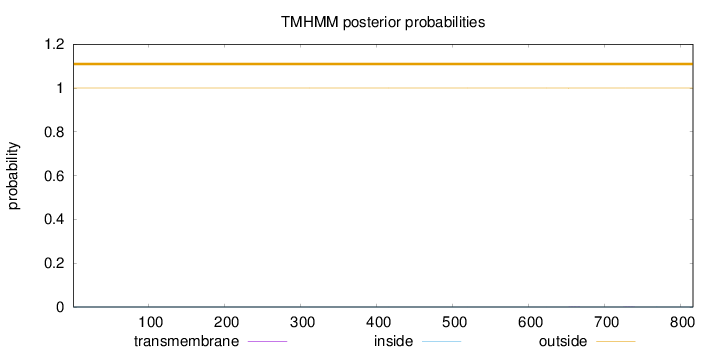
Topology
Length:
816
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00759999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00007
outside
1 - 816
Population Genetic Test Statistics
Pi
26.690753
Theta
186.131277
Tajima's D
-1.747569
CLR
165.75897
CSRT
0.0343482825858707
Interpretation
Possibly Positive selection
