Gene
KWMTBOMO04968 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008085
Annotation
ATP-dependent_RNA_helicase_DDX54_[Papilio_xuthus]
Full name
ATP-dependent RNA helicase
+ More
ATP-dependent RNA helicase DDX54
ATP-dependent RNA helicase DDX54
Location in the cell
Mitochondrial Reliability : 2.389
Sequence
CDS
ATGTTAAAACCTAAAGAATTAGATGATCATCTGCCGGGTTTCGATGCCCCTAAAGCAGATGAGCCTCAAAATGAACCCAAAAACAACCAGAAGAAAAAGAAGAGTTCGGGTGCTTTTCAATCAATGGGCCTAAGTTTTCCTGTTTTAAAAGGAATCACGAAACGGGGTTACAAACAACCAACACCTATACAACGTAAGACCATACCAATAGCATTGACCGGGAAAGATGTTGTTGCAATGGCCCGAACTGGGTCCGGAAAAACAGCTTGTTTTGTCCTCCCTATCCTTGAGAAACTTCTTGTTCCGAACAACAAACCTACACCTGGAAAGAATTTGAGGGCATTAATTCTCTCACCAACTAGAGAACTCGCATTACAGACATTAAGATTTGTGAGGGAGCTAGGAAAATTCACTGGATTAACGAGTGCAGCTATTCTTGGTGGAGAGTCCATTGAGCAGCAGTTTAATGTGATGTCTGGCTCTTCTCCAGACATAGTTGTTGCAACACCTGGGCGCTTCTTACACATCTGCATTGAGATGTGCTTAAAGTTAGACAACATCAAAATAGTTGTTTTTGATGAAGCTGACCGACTATTTGAGCTTGGTTTCGGTGAGCAATTACAAGAGATCTGTGCTCGTCTCCCTTCTAGTCGTCAAACGTTATTATTCTCTGCAACACTACCTAAAATGCTAGTAGAATTTGCGAGGGCTGGACTATCAGATCCCACCTTGATAAGATTAGATGTTGATTGGAAATTACCTTCAACATTATGGCTGGGATGGATATCAGTTAGGGCTGAATTGAAAACGGCAGCCCTGTTGATTCTATTAGACAAAGTATTAACTCCAACAACACCCCAAGCAGTTGTCTTTGCTGCTACGAAACATCATGTTGAGTATTTACATTTGATTCTACAACAAGCCGGTATAAGTTCAACATATGCGTATTCAGGCCTTGATCCGTCAGCCCGTAAGATAGCTCTAGGACGGTTTACCAATAAGAAGTGTTCTGTTCTAATAGTAACGGATGTGGCAGCCAGAGGTCTAGACTTACCTGCATTGGATACAGTCATTAATTACAACTTTCCGGCGAAACCCAAACTCTTCGTCCACAGAGTTGGTCGCAGTGCCAGAGCGGGTCGAGCGGGTCGGGCCTTCTCCCTGGTGGCTGCGGAAGACGTGGCCCACTTGTTGGATCTGCAGCTGTTCTTGGGCACCACGCTGGGGAGAGTGGACGAGAGCGCGGGGCGGGGGGCCGCCTGCGCCAGCGGAGTGTGGGGGGCGTTGCCCGCAGAACAGCTGGAACTTCGACATCAGGACGTTATGGCCTGGGAGAAGAACTACTCTGAAATTGAGGAGGCGGCTCGTACGTGCGCCCGTGGACTGCAGCAGTACGTGCGGTGGCGCGAGGCGGCCTCCGCGGAGGGCAACAGGCGCGCCAAGCTGACCGAAGCGCCCACGCAGCCGCACCCCTTCCTCACCGACGGCGCCCCGGACTCCGCCGCGGTACTCGTGGATCAGATCAAGAACTACACACCTAAAGGGACAATATTGGAGTTGTCGGCTAAAAACGATTCACCGATGTATTTAGCGATGAAAGCCAAGAAGCTCGCACACGGCAGAACTGTCGAGAAGACGAGACAAAACAAAAAGCTACAGGCCGAAGGTAAACTTGATGATATCGTTACTGCCGTTAAATCTAAGAAGAAAAAGAAGGGACCAGTGCGAGATGAGAACTACATACCGCATCAAGCAAGCGACCAGCACACTGAAACAGGCATGGCGGTGAACTTCAGCGCGGGCGCGGAGCACGCCCAGCTGGAGCTGCGAGCCGACAGCGGGGACGCCGAGCGCGCGCAGCGGAACGTCATGCGCTGGGACAGGAAGCGGAAGAAGATGGTGCACGTTGACCCCGACGGCGGACGCAAGATGATTCGTACTGAGAGCGGGTTCCGGGTCCCGGCTTCGTTCCGCAGCGGCCGCTTCGACGACTGGAAAAAACGCAACCTCGCGCACGACCACTCCGACGACGAACCGCAAGCACAAACAAATAAAAGGAAACCAGCGTCGGAGTTCCGTCCGCACTGGGTGAAGCACAACGAACGCGTAGCGAAAAAAGCGGCCGAGTCCAAGTCGCGCGAGTTCAGAGACAAGCAGCAGATCGTCAAGGAGCGCATGCGGAAGGACAAGATTAAACAGAAACTCAAGTTCAAAGGGAACAAGAAGAAAAACAAAAGGAAAAAATAA
Protein
MLKPKELDDHLPGFDAPKADEPQNEPKNNQKKKKSSGAFQSMGLSFPVLKGITKRGYKQPTPIQRKTIPIALTGKDVVAMARTGSGKTACFVLPILEKLLVPNNKPTPGKNLRALILSPTRELALQTLRFVRELGKFTGLTSAAILGGESIEQQFNVMSGSSPDIVVATPGRFLHICIEMCLKLDNIKIVVFDEADRLFELGFGEQLQEICARLPSSRQTLLFSATLPKMLVEFARAGLSDPTLIRLDVDWKLPSTLWLGWISVRAELKTAALLILLDKVLTPTTPQAVVFAATKHHVEYLHLILQQAGISSTYAYSGLDPSARKIALGRFTNKKCSVLIVTDVAARGLDLPALDTVINYNFPAKPKLFVHRVGRSARAGRAGRAFSLVAAEDVAHLLDLQLFLGTTLGRVDESAGRGAACASGVWGALPAEQLELRHQDVMAWEKNYSEIEEAARTCARGLQQYVRWREAASAEGNRRAKLTEAPTQPHPFLTDGAPDSAAVLVDQIKNYTPKGTILELSAKNDSPMYLAMKAKKLAHGRTVEKTRQNKKLQAEGKLDDIVTAVKSKKKKKGPVRDENYIPHQASDQHTETGMAVNFSAGAEHAQLELRADSGDAERAQRNVMRWDRKRKKMVHVDPDGGRKMIRTESGFRVPASFRSGRFDDWKKRNLAHDHSDDEPQAQTNKRKPASEFRPHWVKHNERVAKKAAESKSREFRDKQQIVKERMRKDKIKQKLKFKGNKKKNKRKK
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family. DDX54/DBP10 subfamily.
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A2W1BW47
A0A2A4J9A3
A0A194QE00
A0A2H1VTB7
A0A212FLP5
S4PEY8
+ More
A0A194QKB4 A0A3S2N623 A0A2A4JAU0 A0A195DZ19 A0A195ASZ0 A0A195CN85 A0A195FIN9 A0A158P3U4 E9J1K7 A0A151X1D7 A0A2J7PF04 A0A1Y1LB33 A0A026W6R2 F4WKY0 A0A0M9A3H3 A0A087ZPK1 A0A1B6FMG5 A0A2A3EUU4 A0A1B6L8V3 A0A154P1S1 E2AM45 E0VQ55 A0A023F200 A0A0J7L8J3 A0A0C9R8M7 K7J2Z7 T1I047 A0A0V0G866 A0A139WB42 A0A0L7QZN4 A0A232FIL8 A0A069DW61 A0A224XKB7 A0A1B6CB73 E2C7I6 U4U281 A0A1I8NL41 A0A1I8Q710 A0A1I8Q6Y7 A0A1S3HBS9 A0A0K8T610 Q8SY39 A0A0A9WEU9 B3NEP1 B4QEZ2 A0A226EK89 A7RST4 A0A1J1J9R9 B4HVF1 A0A0K2TUK3 B4K7A0 B3M8Z0 N6UFA2 B4PC53 A0A2H8TQY9 A0A2S2PPP4 J9JS39 A0A1B0CH93 A0A2S2QE53 A0A293MXS0 A0A1B0BRA4 D3ZHY6 A0A1W4Z5J9 H3AKH6 A0A1D1VTK7 A0A1X7ULV9 A0A224YS12 H0WCG4 A0A250YLI6 A0A0P7TFZ0 W5MPL9 A0A131XC94 A0A151N5A2 A0A1U7RF51 A0A3L8SCF7 U3JZ66 M3ZT06 A0A2I4CVV7 A0A1A6FZ34 A0A2Y9KNP7 A0A1D2NBX4 A0A218V3H5 A0A3B4CD54 A0A3Q2PSF5 A0A1V4JSL5 U3CWT5 V3ZA05 A0A3P4S3V6 V9KC54 U3DSV4 A0A146N091
A0A194QKB4 A0A3S2N623 A0A2A4JAU0 A0A195DZ19 A0A195ASZ0 A0A195CN85 A0A195FIN9 A0A158P3U4 E9J1K7 A0A151X1D7 A0A2J7PF04 A0A1Y1LB33 A0A026W6R2 F4WKY0 A0A0M9A3H3 A0A087ZPK1 A0A1B6FMG5 A0A2A3EUU4 A0A1B6L8V3 A0A154P1S1 E2AM45 E0VQ55 A0A023F200 A0A0J7L8J3 A0A0C9R8M7 K7J2Z7 T1I047 A0A0V0G866 A0A139WB42 A0A0L7QZN4 A0A232FIL8 A0A069DW61 A0A224XKB7 A0A1B6CB73 E2C7I6 U4U281 A0A1I8NL41 A0A1I8Q710 A0A1I8Q6Y7 A0A1S3HBS9 A0A0K8T610 Q8SY39 A0A0A9WEU9 B3NEP1 B4QEZ2 A0A226EK89 A7RST4 A0A1J1J9R9 B4HVF1 A0A0K2TUK3 B4K7A0 B3M8Z0 N6UFA2 B4PC53 A0A2H8TQY9 A0A2S2PPP4 J9JS39 A0A1B0CH93 A0A2S2QE53 A0A293MXS0 A0A1B0BRA4 D3ZHY6 A0A1W4Z5J9 H3AKH6 A0A1D1VTK7 A0A1X7ULV9 A0A224YS12 H0WCG4 A0A250YLI6 A0A0P7TFZ0 W5MPL9 A0A131XC94 A0A151N5A2 A0A1U7RF51 A0A3L8SCF7 U3JZ66 M3ZT06 A0A2I4CVV7 A0A1A6FZ34 A0A2Y9KNP7 A0A1D2NBX4 A0A218V3H5 A0A3B4CD54 A0A3Q2PSF5 A0A1V4JSL5 U3CWT5 V3ZA05 A0A3P4S3V6 V9KC54 U3DSV4 A0A146N091
EC Number
3.6.4.13
Pubmed
28756777
26354079
22118469
23622113
21347285
21282665
+ More
28004739 24508170 30249741 21719571 20798317 20566863 25474469 20075255 18362917 19820115 28648823 26334808 23537049 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 17994087 22936249 17615350 17550304 15057822 22673903 9215903 27649274 28797301 21993624 28087693 28049606 22293439 30282656 23542700 27289101 25243066 23254933 24402279
28004739 24508170 30249741 21719571 20798317 20566863 25474469 20075255 18362917 19820115 28648823 26334808 23537049 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 17994087 22936249 17615350 17550304 15057822 22673903 9215903 27649274 28797301 21993624 28087693 28049606 22293439 30282656 23542700 27289101 25243066 23254933 24402279
EMBL
KZ149974
PZC75973.1
NWSH01002321
PCG68531.1
KQ459144
KPJ03702.1
+ More
ODYU01004324 SOQ44070.1 AGBW02007748 OWR54619.1 GAIX01004282 JAA88278.1 KQ461198 KPJ06013.1 RSAL01000337 RVE42440.1 PCG68532.1 KQ980044 KYN18076.1 KQ976746 KYM75333.1 KQ977513 KYN02211.1 KQ981523 KYN40530.1 ADTU01008551 GL767674 EFZ13254.1 KQ982601 KYQ54001.1 NEVH01026086 PNF14916.1 GEZM01062667 GEZM01062666 JAV69550.1 KK107373 QOIP01000004 EZA51658.1 RLU23414.1 GL888206 EGI65222.1 KQ435774 KOX75099.1 GECZ01018415 JAS51354.1 KZ288186 PBC34831.1 GEBQ01019936 JAT20041.1 KQ434787 KZC05284.1 GL440704 EFN65503.1 DS235389 EEB15511.1 GBBI01003120 JAC15592.1 LBMM01000206 KMR04491.1 GBYB01003033 GBYB01004480 JAG72800.1 JAG74247.1 ACPB03019371 GECL01001793 JAP04331.1 KQ971374 KYB25129.1 KQ414679 KOC64058.1 NNAY01000174 OXU30308.1 GBGD01000586 JAC88303.1 GFTR01007945 JAW08481.1 GEDC01026571 GEDC01008684 JAS10727.1 JAS28614.1 GL453383 EFN76062.1 KB631815 ERL86393.1 GBRD01004965 JAG60856.1 AY075397 AE014296 AAL68229.1 AAN11439.1 GBHO01038583 GBHO01038581 JAG05021.1 JAG05023.1 CH954178 EDV50036.1 CM000362 CM002912 EDX07019.1 KMY96638.1 LNIX01000003 OXA57618.1 DS469535 EDO45469.1 CVRI01000075 CRL08620.1 CH480817 EDW49916.1 HACA01011725 CDW29086.1 CH933806 EDW14224.1 CH902618 EDV38934.1 APGK01029188 KB740727 ENN79306.1 CM000159 EDW92708.2 GFXV01004802 MBW16607.1 GGMR01018821 MBY31440.1 ABLF02027827 AJWK01012118 AJWK01012119 GGMS01006823 MBY76026.1 GFWV01020875 MAA45603.1 JXJN01019010 AABR07036387 AFYH01161647 AFYH01161648 AFYH01161649 AFYH01161650 AFYH01161651 AFYH01161652 AFYH01161653 AFYH01161654 AFYH01161655 BDGG01000009 GAV03543.1 GFPF01006235 MAA17381.1 AAKN02056159 GFFW01000285 JAV44503.1 JARO02011672 KPP59736.1 AHAT01005287 AHAT01005288 GEFH01004589 JAP63992.1 AKHW03004053 KYO31779.1 QUSF01000030 RLV99955.1 AGTO01000187 LZPO01110310 OBS58825.1 LJIJ01000096 ODN02763.1 MUZQ01000064 OWK60260.1 LSYS01006629 OPJ74707.1 GAMR01009965 GAMP01003988 JAB23967.1 JAB48767.1 KB202823 ESO87808.1 CYRY02046229 VCX41884.1 JW862960 AFO95477.1 GAMT01004132 GAMS01005654 GAMS01005652 GAMQ01000068 JAB07729.1 JAB17482.1 JAB41783.1 GCES01161866 JAQ24456.1
ODYU01004324 SOQ44070.1 AGBW02007748 OWR54619.1 GAIX01004282 JAA88278.1 KQ461198 KPJ06013.1 RSAL01000337 RVE42440.1 PCG68532.1 KQ980044 KYN18076.1 KQ976746 KYM75333.1 KQ977513 KYN02211.1 KQ981523 KYN40530.1 ADTU01008551 GL767674 EFZ13254.1 KQ982601 KYQ54001.1 NEVH01026086 PNF14916.1 GEZM01062667 GEZM01062666 JAV69550.1 KK107373 QOIP01000004 EZA51658.1 RLU23414.1 GL888206 EGI65222.1 KQ435774 KOX75099.1 GECZ01018415 JAS51354.1 KZ288186 PBC34831.1 GEBQ01019936 JAT20041.1 KQ434787 KZC05284.1 GL440704 EFN65503.1 DS235389 EEB15511.1 GBBI01003120 JAC15592.1 LBMM01000206 KMR04491.1 GBYB01003033 GBYB01004480 JAG72800.1 JAG74247.1 ACPB03019371 GECL01001793 JAP04331.1 KQ971374 KYB25129.1 KQ414679 KOC64058.1 NNAY01000174 OXU30308.1 GBGD01000586 JAC88303.1 GFTR01007945 JAW08481.1 GEDC01026571 GEDC01008684 JAS10727.1 JAS28614.1 GL453383 EFN76062.1 KB631815 ERL86393.1 GBRD01004965 JAG60856.1 AY075397 AE014296 AAL68229.1 AAN11439.1 GBHO01038583 GBHO01038581 JAG05021.1 JAG05023.1 CH954178 EDV50036.1 CM000362 CM002912 EDX07019.1 KMY96638.1 LNIX01000003 OXA57618.1 DS469535 EDO45469.1 CVRI01000075 CRL08620.1 CH480817 EDW49916.1 HACA01011725 CDW29086.1 CH933806 EDW14224.1 CH902618 EDV38934.1 APGK01029188 KB740727 ENN79306.1 CM000159 EDW92708.2 GFXV01004802 MBW16607.1 GGMR01018821 MBY31440.1 ABLF02027827 AJWK01012118 AJWK01012119 GGMS01006823 MBY76026.1 GFWV01020875 MAA45603.1 JXJN01019010 AABR07036387 AFYH01161647 AFYH01161648 AFYH01161649 AFYH01161650 AFYH01161651 AFYH01161652 AFYH01161653 AFYH01161654 AFYH01161655 BDGG01000009 GAV03543.1 GFPF01006235 MAA17381.1 AAKN02056159 GFFW01000285 JAV44503.1 JARO02011672 KPP59736.1 AHAT01005287 AHAT01005288 GEFH01004589 JAP63992.1 AKHW03004053 KYO31779.1 QUSF01000030 RLV99955.1 AGTO01000187 LZPO01110310 OBS58825.1 LJIJ01000096 ODN02763.1 MUZQ01000064 OWK60260.1 LSYS01006629 OPJ74707.1 GAMR01009965 GAMP01003988 JAB23967.1 JAB48767.1 KB202823 ESO87808.1 CYRY02046229 VCX41884.1 JW862960 AFO95477.1 GAMT01004132 GAMS01005654 GAMS01005652 GAMQ01000068 JAB07729.1 JAB17482.1 JAB41783.1 GCES01161866 JAQ24456.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000283053
UP000078492
+ More
UP000078540 UP000078542 UP000078541 UP000005205 UP000075809 UP000235965 UP000053097 UP000279307 UP000007755 UP000053105 UP000005203 UP000242457 UP000076502 UP000000311 UP000009046 UP000036403 UP000002358 UP000015103 UP000007266 UP000053825 UP000215335 UP000008237 UP000030742 UP000095301 UP000095300 UP000085678 UP000000803 UP000008711 UP000000304 UP000198287 UP000001593 UP000183832 UP000001292 UP000009192 UP000007801 UP000019118 UP000002282 UP000007819 UP000092461 UP000092460 UP000002494 UP000192224 UP000008672 UP000186922 UP000007879 UP000005447 UP000034805 UP000018468 UP000050525 UP000189705 UP000276834 UP000016665 UP000002852 UP000192220 UP000092124 UP000248482 UP000094527 UP000197619 UP000261440 UP000265000 UP000190648 UP000030746
UP000078540 UP000078542 UP000078541 UP000005205 UP000075809 UP000235965 UP000053097 UP000279307 UP000007755 UP000053105 UP000005203 UP000242457 UP000076502 UP000000311 UP000009046 UP000036403 UP000002358 UP000015103 UP000007266 UP000053825 UP000215335 UP000008237 UP000030742 UP000095301 UP000095300 UP000085678 UP000000803 UP000008711 UP000000304 UP000198287 UP000001593 UP000183832 UP000001292 UP000009192 UP000007801 UP000019118 UP000002282 UP000007819 UP000092461 UP000092460 UP000002494 UP000192224 UP000008672 UP000186922 UP000007879 UP000005447 UP000034805 UP000018468 UP000050525 UP000189705 UP000276834 UP000016665 UP000002852 UP000192220 UP000092124 UP000248482 UP000094527 UP000197619 UP000261440 UP000265000 UP000190648 UP000030746
Interpro
IPR014001
Helicase_ATP-bd
+ More
IPR000629 RNA-helicase_DEAD-box_CS
IPR001650 Helicase_C
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR033517 DDX54/DBP10
IPR012541 DBP10_C
IPR027417 P-loop_NTPase
IPR001873 ENaC
IPR011009 Kinase-like_dom_sf
IPR004147 UbiB_dom
IPR000719 Prot_kinase_dom
IPR000629 RNA-helicase_DEAD-box_CS
IPR001650 Helicase_C
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR033517 DDX54/DBP10
IPR012541 DBP10_C
IPR027417 P-loop_NTPase
IPR001873 ENaC
IPR011009 Kinase-like_dom_sf
IPR004147 UbiB_dom
IPR000719 Prot_kinase_dom
CDD
ProteinModelPortal
A0A2W1BW47
A0A2A4J9A3
A0A194QE00
A0A2H1VTB7
A0A212FLP5
S4PEY8
+ More
A0A194QKB4 A0A3S2N623 A0A2A4JAU0 A0A195DZ19 A0A195ASZ0 A0A195CN85 A0A195FIN9 A0A158P3U4 E9J1K7 A0A151X1D7 A0A2J7PF04 A0A1Y1LB33 A0A026W6R2 F4WKY0 A0A0M9A3H3 A0A087ZPK1 A0A1B6FMG5 A0A2A3EUU4 A0A1B6L8V3 A0A154P1S1 E2AM45 E0VQ55 A0A023F200 A0A0J7L8J3 A0A0C9R8M7 K7J2Z7 T1I047 A0A0V0G866 A0A139WB42 A0A0L7QZN4 A0A232FIL8 A0A069DW61 A0A224XKB7 A0A1B6CB73 E2C7I6 U4U281 A0A1I8NL41 A0A1I8Q710 A0A1I8Q6Y7 A0A1S3HBS9 A0A0K8T610 Q8SY39 A0A0A9WEU9 B3NEP1 B4QEZ2 A0A226EK89 A7RST4 A0A1J1J9R9 B4HVF1 A0A0K2TUK3 B4K7A0 B3M8Z0 N6UFA2 B4PC53 A0A2H8TQY9 A0A2S2PPP4 J9JS39 A0A1B0CH93 A0A2S2QE53 A0A293MXS0 A0A1B0BRA4 D3ZHY6 A0A1W4Z5J9 H3AKH6 A0A1D1VTK7 A0A1X7ULV9 A0A224YS12 H0WCG4 A0A250YLI6 A0A0P7TFZ0 W5MPL9 A0A131XC94 A0A151N5A2 A0A1U7RF51 A0A3L8SCF7 U3JZ66 M3ZT06 A0A2I4CVV7 A0A1A6FZ34 A0A2Y9KNP7 A0A1D2NBX4 A0A218V3H5 A0A3B4CD54 A0A3Q2PSF5 A0A1V4JSL5 U3CWT5 V3ZA05 A0A3P4S3V6 V9KC54 U3DSV4 A0A146N091
A0A194QKB4 A0A3S2N623 A0A2A4JAU0 A0A195DZ19 A0A195ASZ0 A0A195CN85 A0A195FIN9 A0A158P3U4 E9J1K7 A0A151X1D7 A0A2J7PF04 A0A1Y1LB33 A0A026W6R2 F4WKY0 A0A0M9A3H3 A0A087ZPK1 A0A1B6FMG5 A0A2A3EUU4 A0A1B6L8V3 A0A154P1S1 E2AM45 E0VQ55 A0A023F200 A0A0J7L8J3 A0A0C9R8M7 K7J2Z7 T1I047 A0A0V0G866 A0A139WB42 A0A0L7QZN4 A0A232FIL8 A0A069DW61 A0A224XKB7 A0A1B6CB73 E2C7I6 U4U281 A0A1I8NL41 A0A1I8Q710 A0A1I8Q6Y7 A0A1S3HBS9 A0A0K8T610 Q8SY39 A0A0A9WEU9 B3NEP1 B4QEZ2 A0A226EK89 A7RST4 A0A1J1J9R9 B4HVF1 A0A0K2TUK3 B4K7A0 B3M8Z0 N6UFA2 B4PC53 A0A2H8TQY9 A0A2S2PPP4 J9JS39 A0A1B0CH93 A0A2S2QE53 A0A293MXS0 A0A1B0BRA4 D3ZHY6 A0A1W4Z5J9 H3AKH6 A0A1D1VTK7 A0A1X7ULV9 A0A224YS12 H0WCG4 A0A250YLI6 A0A0P7TFZ0 W5MPL9 A0A131XC94 A0A151N5A2 A0A1U7RF51 A0A3L8SCF7 U3JZ66 M3ZT06 A0A2I4CVV7 A0A1A6FZ34 A0A2Y9KNP7 A0A1D2NBX4 A0A218V3H5 A0A3B4CD54 A0A3Q2PSF5 A0A1V4JSL5 U3CWT5 V3ZA05 A0A3P4S3V6 V9KC54 U3DSV4 A0A146N091
PDB
5IVL
E-value=4.40298e-52,
Score=520
Ontologies
GO
PANTHER
Topology
Subcellular location
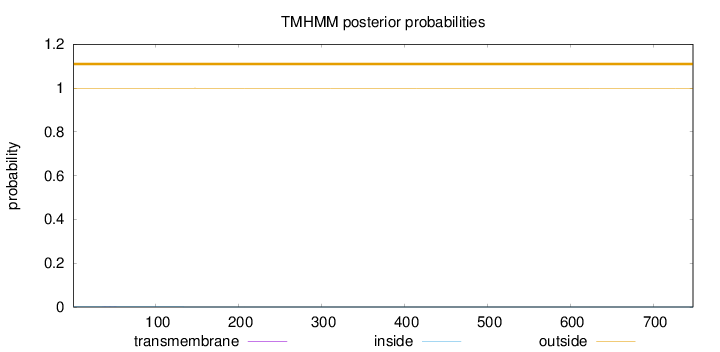
Length:
748
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00678999999999998
Exp number, first 60 AAs:
0.0029
Total prob of N-in:
0.00079
outside
1 - 748
Population Genetic Test Statistics
Pi
191.157738
Theta
160.240009
Tajima's D
-1.720107
CLR
1697.980891
CSRT
0.0369981500924954
Interpretation
Possibly Positive selection
