Gene
KWMTBOMO04967
Pre Gene Modal
BGIBMGA007987
Annotation
peptidoglycan_recognition_protein_S2_[Bombyx_mori]
Full name
Peptidoglycan-recognition protein
+ More
Peptidoglycan recognition protein
Peptidoglycan-recognition protein SA
Peptidoglycan recognition protein 1
Peptidoglycan recognition protein
Peptidoglycan-recognition protein SA
Peptidoglycan recognition protein 1
Alternative Name
Protein semmelweis
Peptidoglycan recognition protein short
Peptidoglycan recognition protein short
Location in the cell
Extracellular Reliability : 3.056
Sequence
CDS
ATGTTGGTTGCACCGTCTCTATTATTTTTGGTGTTCTTGGTGAGTTTCGGAACCCTGAATGCAGCGTCCGAATGCGGCGAAATTCCCATCACCGAATGGAGTGGCACGGAGTCACGTCGTAAACAACCTCTGAAGAGTCCTATTGACTTGGTGGTGATACAACACACGGTATCCAACGATTGCTTTACAGATGAAGAGTGTTTGCTAAGCGTAAATTCTCTACGACAACATCATATGCTTCTGGCTGGGTTCAAGGACTTGGGCTATTCATTCGTGGCTGGAGGCAACGGAAAAATTTATGAAGGAGCGGGATGGAACCATATCGGTGCTCACACATTGCACTACAATAATATATCCATAGGGATCGGTTTCATTGGAGACTTTAGGGAGAAGCTGCCGACCCAGCAGGCACTGCAGGCGGTCCAAGACTTTTTAGCCTGTGGAGTTGAAAATAACTTATTGACTGAAGACTACCACGTCGTTGGTCACCAGCAGTTGATAAACACGCTAAGTCCTGGAGCTGTACTGCAATCAGAAATCGAAAGTTGGCCCCATTGGCTTGATAATGCTCGAAAAGTACTGGGTTAA
Protein
MLVAPSLLFLVFLVSFGTLNAASECGEIPITEWSGTESRRKQPLKSPIDLVVIQHTVSNDCFTDEECLLSVNSLRQHHMLLAGFKDLGYSFVAGGNGKIYEGAGWNHIGAHTLHYNNISIGIGFIGDFREKLPTQQALQAVQDFLACGVENNLLTEDYHVVGHQQLINTLSPGAVLQSEIESWPHWLDNARKVLG
Summary
Description
Binds specifically to peptidoglycan and triggers the propenoloxidase cascade which is an important insect defense mechanism.
Peptidoglycan-recognition protein that plays a key role in innate immunity by binding to peptidoglycans (PGN) of Gram-positive bacteria and activating the Toll pathway upstream of spz activating enzyme SPE (PubMed:11106397, PubMed:16399077, PubMed:15448690). Has no activity against Gram-negative bacteria and fungi (PubMed:11742401). Shows some partial redundancy with PRPGP-SD in Gram-positive bacteria recognition (PubMed:11742401, PubMed:15448690). May act by forming a complex with GNBP1 that activates the proteolytic cleavage of Spatzle and the subsequent activation of Toll pathway (PubMed:14684822, PubMed:14722090, PubMed:11742401). Binds to diaminopimelic acid-type tetrapeptide PGN (DAP-type PGN) and lysine-type PGN (Lys-type PGN) (PubMed:15361936). Has some L,D-carboxypeptidase activity for DAP-type PGN, which are specific to prokaryotes, but not for Lys-type PGN (PubMed:15361936).
Pattern receptor that binds to murein peptidoglycans (PGN) of Gram-positive bacteria. May kill Gram-positive bacteria by interfering with peptidoglycan biosynthesis. Involved in innate immunity.
Pattern receptor that binds to murein peptidoglycans (PGN) of Gram-positive bacteria. Has bactericidal activity towards Gram-positive bacteria. May kill Gram-positive bacteria by interfering with peptidoglycan biosynthesis. Binds also to Gram-negative bacteria, and has bacteriostatic activity towards Gram-negative bacteria. Plays a role in innate immunity.
Peptidoglycan-recognition protein that plays a key role in innate immunity by binding to peptidoglycans (PGN) of Gram-positive bacteria and activating the Toll pathway upstream of spz activating enzyme SPE (PubMed:11106397, PubMed:16399077, PubMed:15448690). Has no activity against Gram-negative bacteria and fungi (PubMed:11742401). Shows some partial redundancy with PRPGP-SD in Gram-positive bacteria recognition (PubMed:11742401, PubMed:15448690). May act by forming a complex with GNBP1 that activates the proteolytic cleavage of Spatzle and the subsequent activation of Toll pathway (PubMed:14684822, PubMed:14722090, PubMed:11742401). Binds to diaminopimelic acid-type tetrapeptide PGN (DAP-type PGN) and lysine-type PGN (Lys-type PGN) (PubMed:15361936). Has some L,D-carboxypeptidase activity for DAP-type PGN, which are specific to prokaryotes, but not for Lys-type PGN (PubMed:15361936).
Pattern receptor that binds to murein peptidoglycans (PGN) of Gram-positive bacteria. May kill Gram-positive bacteria by interfering with peptidoglycan biosynthesis. Involved in innate immunity.
Pattern receptor that binds to murein peptidoglycans (PGN) of Gram-positive bacteria. Has bactericidal activity towards Gram-positive bacteria. May kill Gram-positive bacteria by interfering with peptidoglycan biosynthesis. Binds also to Gram-negative bacteria, and has bacteriostatic activity towards Gram-negative bacteria. Plays a role in innate immunity.
Catalytic Activity
H2O + N-acetyl-D-glucosaminyl-N-acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl-D-alanine = D-alanine + N-acetyl-D-glucosaminyl-N-acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl
Biophysicochemical Properties
21.4 uM for GlcNAc-MurNAc(anhydro)-L-Ala-gamma-D-Glu-meso-DAP-D-Ala
Subunit
Monomer.
Homodimer; disulfide-linked.
Homodimer; disulfide-linked.
Similarity
Belongs to the N-acetylmuramoyl-L-alanine amidase 2 family.
Keywords
Direct protein sequencing
Disulfide bond
Immunity
Innate immunity
Signal
Complete proteome
Reference proteome
3D-structure
Hydrolase
Protease
Secreted
Antibiotic
Antimicrobial
Glycoprotein
Polymorphism
Pyrrolidone carboxylic acid
Feature
chain Peptidoglycan-recognition protein
sequence variant In dbSNP:rs34180629.
sequence variant In dbSNP:rs34180629.
Uniprot
H9JEP0
W6EL05
Q8WSZ1
Q9BLL1
A0A2W1BT81
A0A2H1WXQ3
+ More
A0A346RAC5 A0A212ESF3 A0A194QJS3 A0A194QKT5 E7DN56 B7SFX0 A0A194QR53 W8JAI9 A0A2W1BLP2 O76537 Q0Q026 A0A2A4IX89 B2CZ95 Q8ITT1 A0A212FB31 I4DNW9 A0A194QM25 A0A194QFH8 E5D600 O97369 H9JEU1 Q9XTN0 A0A1Y1M729 Q86RS6 A0A346RAC4 Q86D95 A0A1W4XAW5 A0A2I6B3J1 K7J710 A0A088AJM6 E2BUB1 A0A067QQY1 D7ELY4 A0A0T6ATH0 D2CNK4 A0A1I8NFN3 A0A0U3SX80 A0A3B0KUJ9 A0A0L7KJ86 Q29IN1 A0A125S9Y2 Q0Q025 Q0KKW5 X2JEI8 Q9VYX7 A0A1B0GIX7 A0A1W4UNB6 B4R346 A0A154PIJ2 V5GVK1 A0A1B6GU90 B3NUC2 B4H2R6 B4MAP1 E7DN57 A0A067R4L8 A0A0K8VR92 A0A0M3QYU6 A0A2P8YB58 J3JTK0 Q32S43 B4PY99 A0A067QGE8 A0A2J7Q6X9 V9L8U0 F4WCD8 A0A0C9QKJ8 I3N1R2 A0A067QHE0 A0A1J0M178 B4JXA3 A0A212DE23 F4WMQ9 N6U014 C7AHV4 A0A2R9BV11 H2QGM5 E9J784 B3MY97 A0A158NQ59 A0A0Q9WUD5 A0A0Q5U7H0 H2NZA6 A0A0Q9WW31 A0A2Y9DVG0 T1E303 H8YU85 O75594 V5GS37 A0A1A9Z640 A0A1A9VBQ1 A0A1B0G8B0
A0A346RAC5 A0A212ESF3 A0A194QJS3 A0A194QKT5 E7DN56 B7SFX0 A0A194QR53 W8JAI9 A0A2W1BLP2 O76537 Q0Q026 A0A2A4IX89 B2CZ95 Q8ITT1 A0A212FB31 I4DNW9 A0A194QM25 A0A194QFH8 E5D600 O97369 H9JEU1 Q9XTN0 A0A1Y1M729 Q86RS6 A0A346RAC4 Q86D95 A0A1W4XAW5 A0A2I6B3J1 K7J710 A0A088AJM6 E2BUB1 A0A067QQY1 D7ELY4 A0A0T6ATH0 D2CNK4 A0A1I8NFN3 A0A0U3SX80 A0A3B0KUJ9 A0A0L7KJ86 Q29IN1 A0A125S9Y2 Q0Q025 Q0KKW5 X2JEI8 Q9VYX7 A0A1B0GIX7 A0A1W4UNB6 B4R346 A0A154PIJ2 V5GVK1 A0A1B6GU90 B3NUC2 B4H2R6 B4MAP1 E7DN57 A0A067R4L8 A0A0K8VR92 A0A0M3QYU6 A0A2P8YB58 J3JTK0 Q32S43 B4PY99 A0A067QGE8 A0A2J7Q6X9 V9L8U0 F4WCD8 A0A0C9QKJ8 I3N1R2 A0A067QHE0 A0A1J0M178 B4JXA3 A0A212DE23 F4WMQ9 N6U014 C7AHV4 A0A2R9BV11 H2QGM5 E9J784 B3MY97 A0A158NQ59 A0A0Q9WUD5 A0A0Q5U7H0 H2NZA6 A0A0Q9WW31 A0A2Y9DVG0 T1E303 H8YU85 O75594 V5GS37 A0A1A9Z640 A0A1A9VBQ1 A0A1B0G8B0
EC Number
3.4.17.13
Pubmed
19121390
25797797
28756777
29910977
22118469
26354079
+ More
21167833 9707603 16857061 22651552 9755476 10207004 8662762 28004739 16640722 12225919 20075255 29396863 20798317 24845553 18362917 19820115 25315136 26694822 26227816 15632085 17994087 17434328 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11106397 14738318 12537569 11742401 12032070 14684822 14722090 15448690 16399077 15223330 15361936 29403074 22516182 16269728 17550304 24402279 21719571 29294181 23537049 19387012 22722832 16136131 21282665 21347285 24330624 12975309 15057824 15489334 11461926 16354652 15769462
21167833 9707603 16857061 22651552 9755476 10207004 8662762 28004739 16640722 12225919 20075255 29396863 20798317 24845553 18362917 19820115 25315136 26694822 26227816 15632085 17994087 17434328 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11106397 14738318 12537569 11742401 12032070 14684822 14722090 15448690 16399077 15223330 15361936 29403074 22516182 16269728 17550304 24402279 21719571 29294181 23537049 19387012 22722832 16136131 21282665 21347285 24330624 12975309 15057824 15489334 11461926 16354652 15769462
EMBL
BABH01023631
KF906541
AHJ11178.1
AF441723
AAL32058.1
AB017520
+ More
BAB33295.1 KZ149974 PZC75970.1 ODYU01011865 SOQ57853.1 MH200915 AXS59125.1 AGBW02012798 OWR44425.1 KQ459144 KPJ03701.1 KQ461198 KPJ06009.1 HQ260963 ADU33184.1 EU289210 ABZ81267.1 KPJ06011.1 KF954940 AHK59818.1 PZC75969.1 AF076481 DQ666505 ABG72708.1 NWSH01005638 PCG64086.1 EU557313 ACB45568.1 AF394587 AAN15786.1 AGBW02009391 OWR50928.1 AK403178 BAM19609.1 KPJ06010.1 KPJ03700.1 GU182905 ADR51143.1 AF035445 AAD02029.1 BABH01023291 AB016605 AB016249 GEZM01038634 JAV81582.1 AF413061 AAO21502.1 MH200914 AXS59124.1 AF413068 AAO21509.1 MG570190 AUI41055.1 GL450619 EFN80739.1 KK853478 KDR07310.1 DS497945 EFA12467.1 LJIG01022861 KRT78342.1 EU399240 ACB32179.1 KU218682 ALX00062.1 OUUW01000021 SPP89506.1 JTDY01009650 KOB63145.1 CH379063 EAL32622.2 KT725231 AME17978.1 DQ666506 ABG72709.1 AB232695 BAF03522.1 AE014298 AHN59598.1 AF207540 AF207541 AJ556551 AJ556552 AJ556553 AJ556554 AJ556555 AJ556556 AJ556557 AJ556558 AJ556559 AJ556560 AJ556561 AY075293 AJWK01017804 AJWK01017805 CM000366 EDX17648.1 KQ434924 KZC11632.1 GALX01002754 JAB65712.1 GECZ01003856 JAS65913.1 CH954180 EDV46037.2 CH479204 EDW30633.1 CH940655 EDW66300.1 HQ260964 ADU33185.1 KK852704 KDR18042.1 GDHF01010961 JAI41353.1 CP012528 ALC48229.1 PYGN01000736 PSN41492.1 BT126557 AEE61521.1 AY956814 AAY27976.1 CM000162 EDX01961.1 KDR07311.1 NEVH01017450 PNF24335.1 JW875796 AFP08313.1 GL888070 EGI68118.1 GBYB01004084 JAG73851.1 AGTP01097044 KK853387 KDR07939.1 KX342009 APD13821.1 CH916376 EDV95379.1 MKHE01000004 OWK16479.1 GL888218 EGI64536.1 APGK01054672 APGK01054673 KB741251 ENN71867.1 FJ546159 ACT66863.1 AJFE02059320 AACZ04019598 NBAG03000399 PNI28802.1 GL768424 EFZ11322.1 CH902632 EDV32591.1 ADTU01022971 CH940647 KRF84516.1 CH954178 KQS43684.1 ABGA01274545 ABGA01400468 ABGA01400469 NDHI03003467 PNJ40971.1 KRF84512.1 GALA01000820 JAA94032.1 JF915527 AFD54029.1 AF076483 AF242517 AY358936 AC007785 BC096154 BC096155 BC096156 BC096157 BC101845 BC101847 GALX01005413 JAB63053.1 CCAG010000228
BAB33295.1 KZ149974 PZC75970.1 ODYU01011865 SOQ57853.1 MH200915 AXS59125.1 AGBW02012798 OWR44425.1 KQ459144 KPJ03701.1 KQ461198 KPJ06009.1 HQ260963 ADU33184.1 EU289210 ABZ81267.1 KPJ06011.1 KF954940 AHK59818.1 PZC75969.1 AF076481 DQ666505 ABG72708.1 NWSH01005638 PCG64086.1 EU557313 ACB45568.1 AF394587 AAN15786.1 AGBW02009391 OWR50928.1 AK403178 BAM19609.1 KPJ06010.1 KPJ03700.1 GU182905 ADR51143.1 AF035445 AAD02029.1 BABH01023291 AB016605 AB016249 GEZM01038634 JAV81582.1 AF413061 AAO21502.1 MH200914 AXS59124.1 AF413068 AAO21509.1 MG570190 AUI41055.1 GL450619 EFN80739.1 KK853478 KDR07310.1 DS497945 EFA12467.1 LJIG01022861 KRT78342.1 EU399240 ACB32179.1 KU218682 ALX00062.1 OUUW01000021 SPP89506.1 JTDY01009650 KOB63145.1 CH379063 EAL32622.2 KT725231 AME17978.1 DQ666506 ABG72709.1 AB232695 BAF03522.1 AE014298 AHN59598.1 AF207540 AF207541 AJ556551 AJ556552 AJ556553 AJ556554 AJ556555 AJ556556 AJ556557 AJ556558 AJ556559 AJ556560 AJ556561 AY075293 AJWK01017804 AJWK01017805 CM000366 EDX17648.1 KQ434924 KZC11632.1 GALX01002754 JAB65712.1 GECZ01003856 JAS65913.1 CH954180 EDV46037.2 CH479204 EDW30633.1 CH940655 EDW66300.1 HQ260964 ADU33185.1 KK852704 KDR18042.1 GDHF01010961 JAI41353.1 CP012528 ALC48229.1 PYGN01000736 PSN41492.1 BT126557 AEE61521.1 AY956814 AAY27976.1 CM000162 EDX01961.1 KDR07311.1 NEVH01017450 PNF24335.1 JW875796 AFP08313.1 GL888070 EGI68118.1 GBYB01004084 JAG73851.1 AGTP01097044 KK853387 KDR07939.1 KX342009 APD13821.1 CH916376 EDV95379.1 MKHE01000004 OWK16479.1 GL888218 EGI64536.1 APGK01054672 APGK01054673 KB741251 ENN71867.1 FJ546159 ACT66863.1 AJFE02059320 AACZ04019598 NBAG03000399 PNI28802.1 GL768424 EFZ11322.1 CH902632 EDV32591.1 ADTU01022971 CH940647 KRF84516.1 CH954178 KQS43684.1 ABGA01274545 ABGA01400468 ABGA01400469 NDHI03003467 PNJ40971.1 KRF84512.1 GALA01000820 JAA94032.1 JF915527 AFD54029.1 AF076483 AF242517 AY358936 AC007785 BC096154 BC096155 BC096156 BC096157 BC101845 BC101847 GALX01005413 JAB63053.1 CCAG010000228
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000192223
+ More
UP000002358 UP000005203 UP000008237 UP000027135 UP000007266 UP000095301 UP000268350 UP000037510 UP000001819 UP000000803 UP000092461 UP000192221 UP000000304 UP000076502 UP000008711 UP000008744 UP000008792 UP000092553 UP000245037 UP000002282 UP000235965 UP000007755 UP000005215 UP000001070 UP000019118 UP000240080 UP000002277 UP000007801 UP000005205 UP000001595 UP000248480 UP000005640 UP000092445 UP000078200 UP000092444
UP000002358 UP000005203 UP000008237 UP000027135 UP000007266 UP000095301 UP000268350 UP000037510 UP000001819 UP000000803 UP000092461 UP000192221 UP000000304 UP000076502 UP000008711 UP000008744 UP000008792 UP000092553 UP000245037 UP000002282 UP000235965 UP000007755 UP000005215 UP000001070 UP000019118 UP000240080 UP000002277 UP000007801 UP000005205 UP000001595 UP000248480 UP000005640 UP000092445 UP000078200 UP000092444
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEP0
W6EL05
Q8WSZ1
Q9BLL1
A0A2W1BT81
A0A2H1WXQ3
+ More
A0A346RAC5 A0A212ESF3 A0A194QJS3 A0A194QKT5 E7DN56 B7SFX0 A0A194QR53 W8JAI9 A0A2W1BLP2 O76537 Q0Q026 A0A2A4IX89 B2CZ95 Q8ITT1 A0A212FB31 I4DNW9 A0A194QM25 A0A194QFH8 E5D600 O97369 H9JEU1 Q9XTN0 A0A1Y1M729 Q86RS6 A0A346RAC4 Q86D95 A0A1W4XAW5 A0A2I6B3J1 K7J710 A0A088AJM6 E2BUB1 A0A067QQY1 D7ELY4 A0A0T6ATH0 D2CNK4 A0A1I8NFN3 A0A0U3SX80 A0A3B0KUJ9 A0A0L7KJ86 Q29IN1 A0A125S9Y2 Q0Q025 Q0KKW5 X2JEI8 Q9VYX7 A0A1B0GIX7 A0A1W4UNB6 B4R346 A0A154PIJ2 V5GVK1 A0A1B6GU90 B3NUC2 B4H2R6 B4MAP1 E7DN57 A0A067R4L8 A0A0K8VR92 A0A0M3QYU6 A0A2P8YB58 J3JTK0 Q32S43 B4PY99 A0A067QGE8 A0A2J7Q6X9 V9L8U0 F4WCD8 A0A0C9QKJ8 I3N1R2 A0A067QHE0 A0A1J0M178 B4JXA3 A0A212DE23 F4WMQ9 N6U014 C7AHV4 A0A2R9BV11 H2QGM5 E9J784 B3MY97 A0A158NQ59 A0A0Q9WUD5 A0A0Q5U7H0 H2NZA6 A0A0Q9WW31 A0A2Y9DVG0 T1E303 H8YU85 O75594 V5GS37 A0A1A9Z640 A0A1A9VBQ1 A0A1B0G8B0
A0A346RAC5 A0A212ESF3 A0A194QJS3 A0A194QKT5 E7DN56 B7SFX0 A0A194QR53 W8JAI9 A0A2W1BLP2 O76537 Q0Q026 A0A2A4IX89 B2CZ95 Q8ITT1 A0A212FB31 I4DNW9 A0A194QM25 A0A194QFH8 E5D600 O97369 H9JEU1 Q9XTN0 A0A1Y1M729 Q86RS6 A0A346RAC4 Q86D95 A0A1W4XAW5 A0A2I6B3J1 K7J710 A0A088AJM6 E2BUB1 A0A067QQY1 D7ELY4 A0A0T6ATH0 D2CNK4 A0A1I8NFN3 A0A0U3SX80 A0A3B0KUJ9 A0A0L7KJ86 Q29IN1 A0A125S9Y2 Q0Q025 Q0KKW5 X2JEI8 Q9VYX7 A0A1B0GIX7 A0A1W4UNB6 B4R346 A0A154PIJ2 V5GVK1 A0A1B6GU90 B3NUC2 B4H2R6 B4MAP1 E7DN57 A0A067R4L8 A0A0K8VR92 A0A0M3QYU6 A0A2P8YB58 J3JTK0 Q32S43 B4PY99 A0A067QGE8 A0A2J7Q6X9 V9L8U0 F4WCD8 A0A0C9QKJ8 I3N1R2 A0A067QHE0 A0A1J0M178 B4JXA3 A0A212DE23 F4WMQ9 N6U014 C7AHV4 A0A2R9BV11 H2QGM5 E9J784 B3MY97 A0A158NQ59 A0A0Q9WUD5 A0A0Q5U7H0 H2NZA6 A0A0Q9WW31 A0A2Y9DVG0 T1E303 H8YU85 O75594 V5GS37 A0A1A9Z640 A0A1A9VBQ1 A0A1B0G8B0
PDB
5XZ4
E-value=4.57374e-37,
Score=384
Ontologies
GO
GO:0008270
GO:0009253
GO:0045087
GO:0042834
GO:0008745
GO:0016021
GO:0032827
GO:0050830
GO:0019730
GO:0016019
GO:0005615
GO:0005576
GO:0032500
GO:0045752
GO:0038187
GO:0008329
GO:0008233
GO:0006965
GO:0032494
GO:0061844
GO:0031640
GO:0016045
GO:0051714
GO:0003779
GO:0032689
GO:0070062
GO:1904724
GO:0097013
GO:0043312
GO:0044117
GO:0050728
GO:0035580
GO:0006796
GO:0015074
GO:0007166
GO:0006032
GO:0005515
GO:0016742
GO:0003824
PANTHER
Topology
Subcellular location
Secreted
Cytoplasmic granule
Cytoplasmic granule
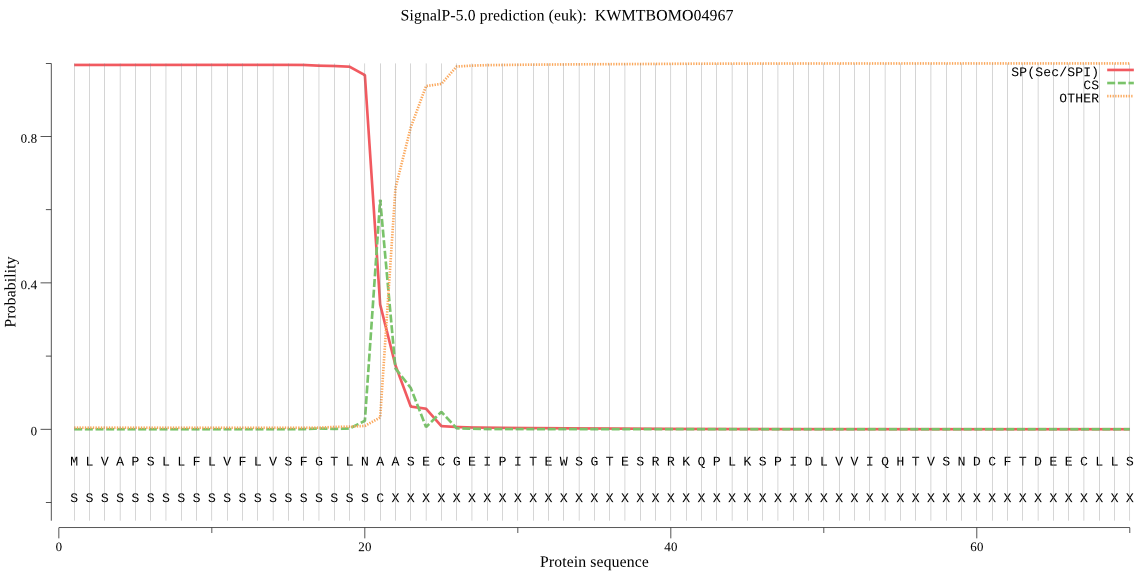
SignalP
Position: 1 - 21,
Likelihood: 0.995534
Length:
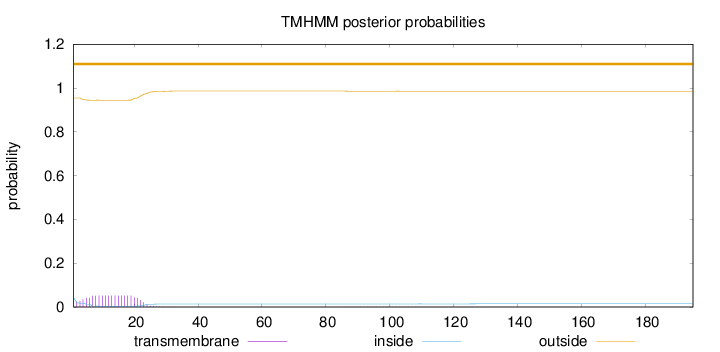
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.03786
Exp number, first 60 AAs:
1.01471
Total prob of N-in:
0.04533
outside
1 - 195
Population Genetic Test Statistics
Pi
316.150662
Theta
192.47319
Tajima's D
1.988019
CLR
0.222576
CSRT
0.877706114694265
Interpretation
Uncertain
