Gene
KWMTBOMO04966 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007988
Annotation
inorganic_pyrophosphatase_[Bombyx_mori]
Full name
Inorganic pyrophosphatase
Alternative Name
Nucleosome-remodeling factor 38 kDa subunit
Pyrophosphate phospho-hydrolase
Pyrophosphate phospho-hydrolase
Location in the cell
Cytoplasmic Reliability : 1.822
Sequence
CDS
ATGTCACTTTCACGTTGCGTTTGTGCCGCAGCTGTTCCGGTTTGCCGGTCGATAGCAAGACGATTGTGCGCTGTTAAAGAACCGACACGTGTAACTTGTTCAATTAATTCGACTGCCACACTAAAAACTCAAGTAAGGATGTACATCGTAGAAGAAAGAGGATCACCTTACACTCCTGACTATCGTGTATTCTTCAAGGATGAAGGCGGCCCTATATCGCCCATGCACGACATTCCACTATGGGCCGACAAAGCTCAACGCCTCGTCAACATGGTAGTAGAAGTACCTAGATGGACCAATGCGAAAATGGAGATCAGCCTCGGGGAGGCCCTCAATCCTATCAAGCAGGACGTAAAGAAAGGCAACCTTCGGTTCGTGAACAACGTCTTCCCTCATCGCGGCTACATCTGGAATTACGGTGCCCTGCCGCAGACCTGGGAGAATCCTAATCACGTCGACCCTGACACGGGCGCGAGGGGCGACAACGACCCGGTTGACGTCATCGAAATCGGTGAGCGCGTCGCGAGTCGCGGTGACGTTTACCCCGTAAAGATCCTCGGTACGCTAGCTCTCATCGACGAAGGCGAGACCGATTGGAAGCTGATAGCGATCGATTCCCGGGACCCGAACGCGGAGAAGTTGAACGATGTACAAGACGTGGAGACGTTGTTCCCTGGCCTCCTCCGCGCCACCGTCGAGTGGTTCAGACTCTACAAAGTTCCGGACGGCAAGCCGGTGAACAAGTTCGCGTTTGACGGCGAAGCTAAGAACGCAGAGTTCGCTTACAAAGTCGTTGACGAGGTTCACGAATTTTGGAAGAGTCTCATCAGCGGCAACGTTCCTGACGCAACCGATATAAACAAAATGAACGTGTCAGTGAGCGGCAGCGTGGACCGGGCGGAGCGCTCCGAAGCCGCCGCGGTGTTGGCGAAGGCTCCAACGCTCGGGCAGCCCGCCGCCATACCGCAGACAGTTGACAAGTGGCACTTCCTCGCGAACCTATAG
Protein
MSLSRCVCAAAVPVCRSIARRLCAVKEPTRVTCSINSTATLKTQVRMYIVEERGSPYTPDYRVFFKDEGGPISPMHDIPLWADKAQRLVNMVVEVPRWTNAKMEISLGEALNPIKQDVKKGNLRFVNNVFPHRGYIWNYGALPQTWENPNHVDPDTGARGDNDPVDVIEIGERVASRGDVYPVKILGTLALIDEGETDWKLIAIDSRDPNAEKLNDVQDVETLFPGLLRATVEWFRLYKVPDGKPVNKFAFDGEAKNAEFAYKVVDEVHEFWKSLISGNVPDATDINKMNVSVSGSVDRAERSEAAAVLAKAPTLGQPAAIPQTVDKWHFLANL
Summary
Description
Component of NURF (nucleosome remodeling factor), a complex which catalyzes ATP-dependent nucleosome sliding and facilitates transcription of chromatin. NURF is required for homeotic gene expression, proper larval blood cell development, normal male X chromosome morphology, ecdysteroid signaling and metamorphosis. Nurf-38 may have adapted to deliver pyrophosphatase to chromatin to assist in replication or transcription by efficient removal of the inhibitory metabolite.
Catalytic Activity
diphosphate + H2O = H(+) + 2 phosphate
Cofactor
Mg(2+)
Subunit
Component of the NURF complex composed of Caf1, E(bx), Nurf-38 and Iswi.
Miscellaneous
The ATPase activity of NURF is stimulated by the presence of nucleosomes rather than by free DNA.
Similarity
Belongs to the PPase family.
Keywords
Chromatin regulator
Complete proteome
Cytoplasm
Direct protein sequencing
Hydrolase
Magnesium
Metal-binding
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Inorganic pyrophosphatase
Uniprot
E5EVW5
A0A2W1B129
A0A2A4IV62
A0A212F0F5
A0A3S2L1J1
A0A194QK65
+ More
A0A0N0P9B8 D6WB60 V5G7A2 A0A336M2U3 A0A182MP03 A0A0L7L7W2 A0A084W7D2 A0A1L8E0P8 A0A1L8E0T9 A0A2M4A8Q7 A0A2M4A849 J3JVA3 A0A182QVM7 A0A2J7RIE7 W5J4U0 A0A2M4BRS6 A0A2M4BRE3 A0A1Q3FY31 A0A2M4CZK7 A0A2J7RID2 A0A2M4CZJ7 U5ES97 A0A2M4CZG4 A0A2M3Z2B5 A0A182UM70 A0A2M4CZH0 A0A182NKT6 A0A182PS37 A0A182XK84 A0A1S4F3Z8 B5E1A1 B4MP06 A0A182W0L8 A0A023ER17 N6T450 Q1HR16 A0A1W4W0N7 Q5TU96 A0A182IP54 A0A182UF49 A0A182HJQ4 E0W2T1 A0A1S4F3X2 Q17G61 A0A182K237 A0A1Y1LHG1 A0A182KWW5 F5HK08 F5HK09 A0A347ZJC1 A0A182G395 A0A067R4J7 F5HK07 Q17G60 T1DPX3 A0A3R7PKM8 A0A0M4ETX4 E9HGN3 A0A0T6B569 A0A162QQ82 A0A0P5AHB4 T1IWU8 A0A0P5QV05 A0A0N8E9X5 A0A0P5N2N8 A0A0N7ZYY7 A0A0P6DIU3 A0A1L8EI58 B4PBC5 A0A3B0J073 A0A1L8EIJ7 A0A1L8EEH8 A0A1L8EBC4 A0A1J1HQT6 A0A1L8EC30 A0A0P6FH14 A0A0M8ZR40 A0A0N7ZHH9 A0A0P5YG10 A0A1Y1LIZ8 I0DHK6 O77460 A0A182SHG8 A0A0N8AX26 A0A2J7RID6 A0A0P6FBJ6 A0A0P6ER08 A0A0J9UAG2 B4KQ66 A0A182Y4J5 B3NQM0 A0A1I8M9Y3 A0A1I8P9N4 B4QC15
A0A0N0P9B8 D6WB60 V5G7A2 A0A336M2U3 A0A182MP03 A0A0L7L7W2 A0A084W7D2 A0A1L8E0P8 A0A1L8E0T9 A0A2M4A8Q7 A0A2M4A849 J3JVA3 A0A182QVM7 A0A2J7RIE7 W5J4U0 A0A2M4BRS6 A0A2M4BRE3 A0A1Q3FY31 A0A2M4CZK7 A0A2J7RID2 A0A2M4CZJ7 U5ES97 A0A2M4CZG4 A0A2M3Z2B5 A0A182UM70 A0A2M4CZH0 A0A182NKT6 A0A182PS37 A0A182XK84 A0A1S4F3Z8 B5E1A1 B4MP06 A0A182W0L8 A0A023ER17 N6T450 Q1HR16 A0A1W4W0N7 Q5TU96 A0A182IP54 A0A182UF49 A0A182HJQ4 E0W2T1 A0A1S4F3X2 Q17G61 A0A182K237 A0A1Y1LHG1 A0A182KWW5 F5HK08 F5HK09 A0A347ZJC1 A0A182G395 A0A067R4J7 F5HK07 Q17G60 T1DPX3 A0A3R7PKM8 A0A0M4ETX4 E9HGN3 A0A0T6B569 A0A162QQ82 A0A0P5AHB4 T1IWU8 A0A0P5QV05 A0A0N8E9X5 A0A0P5N2N8 A0A0N7ZYY7 A0A0P6DIU3 A0A1L8EI58 B4PBC5 A0A3B0J073 A0A1L8EIJ7 A0A1L8EEH8 A0A1L8EBC4 A0A1J1HQT6 A0A1L8EC30 A0A0P6FH14 A0A0M8ZR40 A0A0N7ZHH9 A0A0P5YG10 A0A1Y1LIZ8 I0DHK6 O77460 A0A182SHG8 A0A0N8AX26 A0A2J7RID6 A0A0P6FBJ6 A0A0P6ER08 A0A0J9UAG2 B4KQ66 A0A182Y4J5 B3NQM0 A0A1I8M9Y3 A0A1I8P9N4 B4QC15
EC Number
3.6.1.1
Pubmed
19121390
21457781
28756777
22118469
26354079
18362917
+ More
19820115 26227816 24438588 22516182 20920257 23761445 15632085 17994087 24945155 23537049 17204158 12364791 14747013 17210077 20566863 17510324 28004739 20966253 26483478 24845553 21292972 17550304 9784495 10731132 12537572 12537569 8521501 22936249 25244985 25315136
19820115 26227816 24438588 22516182 20920257 23761445 15632085 17994087 24945155 23537049 17204158 12364791 14747013 17210077 20566863 17510324 28004739 20966253 26483478 24845553 21292972 17550304 9784495 10731132 12537572 12537569 8521501 22936249 25244985 25315136
EMBL
BABH01023630
BABH01023631
HM012809
ADQ89808.1
KZ150479
PZC70732.1
+ More
NWSH01007242 PCG63030.1 AGBW02011129 OWR47181.1 RSAL01000248 RVE43533.1 KQ461198 KPJ05927.1 KQ459566 KPI99608.1 KQ971311 EEZ98942.2 GALX01002481 JAB65985.1 UFQT01000456 SSX24554.1 AXCM01000643 JTDY01002418 KOB71450.1 ATLV01021211 KE525314 KFB46126.1 GFDF01001817 JAV12267.1 GFDF01001818 JAV12266.1 GGFK01003801 MBW37122.1 GGFK01003653 MBW36974.1 BT127170 AEE62132.1 AXCN02000304 NEVH01003502 PNF40596.1 ADMH02002195 ETN57880.1 GGFJ01006571 MBW55712.1 GGFJ01006516 MBW55657.1 GFDL01002653 JAV32392.1 GGFL01006541 MBW70719.1 PNF40595.1 GGFL01006527 MBW70705.1 GANO01003348 JAB56523.1 GGFL01006525 MBW70703.1 GGFM01001918 MBW22669.1 GGFL01006542 MBW70720.1 CM000071 EDY69489.2 CH963848 EDW73845.2 GAPW01002025 JAC11573.1 APGK01053204 KB741222 KB631665 ENN72408.1 ERL85141.1 DQ440278 ABF18311.1 AAAB01008851 EAL40975.4 AXCP01007741 APCN01002203 DS235879 EEB19937.1 CH477266 EAT45523.1 GEZM01057450 JAV72331.1 EGK96617.1 EGK96620.1 EGK96621.1 FX985689 BBA84427.1 JXUM01042241 JXUM01042242 JXUM01042243 JXUM01042244 KQ561316 KXJ79016.1 KK852898 KDR14150.1 EGK96619.1 EAT45524.1 GAMD01002949 JAA98641.1 QCYY01001832 ROT74936.1 CP012524 ALC40757.1 GL732642 EFX69116.1 LJIG01015964 KRT82023.1 LRGB01000311 KZS19865.1 GDIP01198842 JAJ24560.1 JH431630 GDIQ01116101 JAL35625.1 GDIQ01047468 JAN47269.1 GDIQ01147933 JAL03793.1 GDIP01198841 JAJ24561.1 GDIQ01079907 JAN14830.1 GFDG01000387 JAV18412.1 CM000158 EDW92557.2 OUUW01000001 SPP74065.1 GFDG01000386 JAV18413.1 GFDG01001699 JAV17100.1 GFDG01002884 JAV15915.1 CVRI01000019 CRK90407.1 GFDG01002575 JAV16224.1 GDIQ01047467 JAN47270.1 KQ435944 KOX68266.1 GDIP01244905 JAI78496.1 GDIP01058217 JAM45498.1 GEZM01057451 JAV72330.1 BT133451 AFH89824.1 AF085600 AF085601 AE013599 AY075479 GDIQ01234451 JAK17274.1 PNF40599.1 GDIQ01059071 JAN35666.1 GDIQ01059072 JAN35665.1 CM002911 KMY96370.1 CH933808 EDW09194.2 CH954179 EDV57023.2 CM000362 EDX08570.1
NWSH01007242 PCG63030.1 AGBW02011129 OWR47181.1 RSAL01000248 RVE43533.1 KQ461198 KPJ05927.1 KQ459566 KPI99608.1 KQ971311 EEZ98942.2 GALX01002481 JAB65985.1 UFQT01000456 SSX24554.1 AXCM01000643 JTDY01002418 KOB71450.1 ATLV01021211 KE525314 KFB46126.1 GFDF01001817 JAV12267.1 GFDF01001818 JAV12266.1 GGFK01003801 MBW37122.1 GGFK01003653 MBW36974.1 BT127170 AEE62132.1 AXCN02000304 NEVH01003502 PNF40596.1 ADMH02002195 ETN57880.1 GGFJ01006571 MBW55712.1 GGFJ01006516 MBW55657.1 GFDL01002653 JAV32392.1 GGFL01006541 MBW70719.1 PNF40595.1 GGFL01006527 MBW70705.1 GANO01003348 JAB56523.1 GGFL01006525 MBW70703.1 GGFM01001918 MBW22669.1 GGFL01006542 MBW70720.1 CM000071 EDY69489.2 CH963848 EDW73845.2 GAPW01002025 JAC11573.1 APGK01053204 KB741222 KB631665 ENN72408.1 ERL85141.1 DQ440278 ABF18311.1 AAAB01008851 EAL40975.4 AXCP01007741 APCN01002203 DS235879 EEB19937.1 CH477266 EAT45523.1 GEZM01057450 JAV72331.1 EGK96617.1 EGK96620.1 EGK96621.1 FX985689 BBA84427.1 JXUM01042241 JXUM01042242 JXUM01042243 JXUM01042244 KQ561316 KXJ79016.1 KK852898 KDR14150.1 EGK96619.1 EAT45524.1 GAMD01002949 JAA98641.1 QCYY01001832 ROT74936.1 CP012524 ALC40757.1 GL732642 EFX69116.1 LJIG01015964 KRT82023.1 LRGB01000311 KZS19865.1 GDIP01198842 JAJ24560.1 JH431630 GDIQ01116101 JAL35625.1 GDIQ01047468 JAN47269.1 GDIQ01147933 JAL03793.1 GDIP01198841 JAJ24561.1 GDIQ01079907 JAN14830.1 GFDG01000387 JAV18412.1 CM000158 EDW92557.2 OUUW01000001 SPP74065.1 GFDG01000386 JAV18413.1 GFDG01001699 JAV17100.1 GFDG01002884 JAV15915.1 CVRI01000019 CRK90407.1 GFDG01002575 JAV16224.1 GDIQ01047467 JAN47270.1 KQ435944 KOX68266.1 GDIP01244905 JAI78496.1 GDIP01058217 JAM45498.1 GEZM01057451 JAV72330.1 BT133451 AFH89824.1 AF085600 AF085601 AE013599 AY075479 GDIQ01234451 JAK17274.1 PNF40599.1 GDIQ01059071 JAN35666.1 GDIQ01059072 JAN35665.1 CM002911 KMY96370.1 CH933808 EDW09194.2 CH954179 EDV57023.2 CM000362 EDX08570.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000007266 UP000075883 UP000037510 UP000030765 UP000075886 UP000235965 UP000000673 UP000075903 UP000075884 UP000075885 UP000076407 UP000001819 UP000007798 UP000075920 UP000019118 UP000030742 UP000192221 UP000007062 UP000075880 UP000075902 UP000075840 UP000009046 UP000008820 UP000075881 UP000075882 UP000069940 UP000249989 UP000027135 UP000283509 UP000092553 UP000000305 UP000076858 UP000002282 UP000268350 UP000183832 UP000053105 UP000000803 UP000075901 UP000009192 UP000076408 UP000008711 UP000095301 UP000095300 UP000000304
UP000007266 UP000075883 UP000037510 UP000030765 UP000075886 UP000235965 UP000000673 UP000075903 UP000075884 UP000075885 UP000076407 UP000001819 UP000007798 UP000075920 UP000019118 UP000030742 UP000192221 UP000007062 UP000075880 UP000075902 UP000075840 UP000009046 UP000008820 UP000075881 UP000075882 UP000069940 UP000249989 UP000027135 UP000283509 UP000092553 UP000000305 UP000076858 UP000002282 UP000268350 UP000183832 UP000053105 UP000000803 UP000075901 UP000009192 UP000076408 UP000008711 UP000095301 UP000095300 UP000000304
Pfam
PF00719 Pyrophosphatase
SUPFAM
SSF50324
SSF50324
Gene 3D
ProteinModelPortal
E5EVW5
A0A2W1B129
A0A2A4IV62
A0A212F0F5
A0A3S2L1J1
A0A194QK65
+ More
A0A0N0P9B8 D6WB60 V5G7A2 A0A336M2U3 A0A182MP03 A0A0L7L7W2 A0A084W7D2 A0A1L8E0P8 A0A1L8E0T9 A0A2M4A8Q7 A0A2M4A849 J3JVA3 A0A182QVM7 A0A2J7RIE7 W5J4U0 A0A2M4BRS6 A0A2M4BRE3 A0A1Q3FY31 A0A2M4CZK7 A0A2J7RID2 A0A2M4CZJ7 U5ES97 A0A2M4CZG4 A0A2M3Z2B5 A0A182UM70 A0A2M4CZH0 A0A182NKT6 A0A182PS37 A0A182XK84 A0A1S4F3Z8 B5E1A1 B4MP06 A0A182W0L8 A0A023ER17 N6T450 Q1HR16 A0A1W4W0N7 Q5TU96 A0A182IP54 A0A182UF49 A0A182HJQ4 E0W2T1 A0A1S4F3X2 Q17G61 A0A182K237 A0A1Y1LHG1 A0A182KWW5 F5HK08 F5HK09 A0A347ZJC1 A0A182G395 A0A067R4J7 F5HK07 Q17G60 T1DPX3 A0A3R7PKM8 A0A0M4ETX4 E9HGN3 A0A0T6B569 A0A162QQ82 A0A0P5AHB4 T1IWU8 A0A0P5QV05 A0A0N8E9X5 A0A0P5N2N8 A0A0N7ZYY7 A0A0P6DIU3 A0A1L8EI58 B4PBC5 A0A3B0J073 A0A1L8EIJ7 A0A1L8EEH8 A0A1L8EBC4 A0A1J1HQT6 A0A1L8EC30 A0A0P6FH14 A0A0M8ZR40 A0A0N7ZHH9 A0A0P5YG10 A0A1Y1LIZ8 I0DHK6 O77460 A0A182SHG8 A0A0N8AX26 A0A2J7RID6 A0A0P6FBJ6 A0A0P6ER08 A0A0J9UAG2 B4KQ66 A0A182Y4J5 B3NQM0 A0A1I8M9Y3 A0A1I8P9N4 B4QC15
A0A0N0P9B8 D6WB60 V5G7A2 A0A336M2U3 A0A182MP03 A0A0L7L7W2 A0A084W7D2 A0A1L8E0P8 A0A1L8E0T9 A0A2M4A8Q7 A0A2M4A849 J3JVA3 A0A182QVM7 A0A2J7RIE7 W5J4U0 A0A2M4BRS6 A0A2M4BRE3 A0A1Q3FY31 A0A2M4CZK7 A0A2J7RID2 A0A2M4CZJ7 U5ES97 A0A2M4CZG4 A0A2M3Z2B5 A0A182UM70 A0A2M4CZH0 A0A182NKT6 A0A182PS37 A0A182XK84 A0A1S4F3Z8 B5E1A1 B4MP06 A0A182W0L8 A0A023ER17 N6T450 Q1HR16 A0A1W4W0N7 Q5TU96 A0A182IP54 A0A182UF49 A0A182HJQ4 E0W2T1 A0A1S4F3X2 Q17G61 A0A182K237 A0A1Y1LHG1 A0A182KWW5 F5HK08 F5HK09 A0A347ZJC1 A0A182G395 A0A067R4J7 F5HK07 Q17G60 T1DPX3 A0A3R7PKM8 A0A0M4ETX4 E9HGN3 A0A0T6B569 A0A162QQ82 A0A0P5AHB4 T1IWU8 A0A0P5QV05 A0A0N8E9X5 A0A0P5N2N8 A0A0N7ZYY7 A0A0P6DIU3 A0A1L8EI58 B4PBC5 A0A3B0J073 A0A1L8EIJ7 A0A1L8EEH8 A0A1L8EBC4 A0A1J1HQT6 A0A1L8EC30 A0A0P6FH14 A0A0M8ZR40 A0A0N7ZHH9 A0A0P5YG10 A0A1Y1LIZ8 I0DHK6 O77460 A0A182SHG8 A0A0N8AX26 A0A2J7RID6 A0A0P6FBJ6 A0A0P6ER08 A0A0J9UAG2 B4KQ66 A0A182Y4J5 B3NQM0 A0A1I8M9Y3 A0A1I8P9N4 B4QC15
PDB
2IHP
E-value=5.0733e-89,
Score=835
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
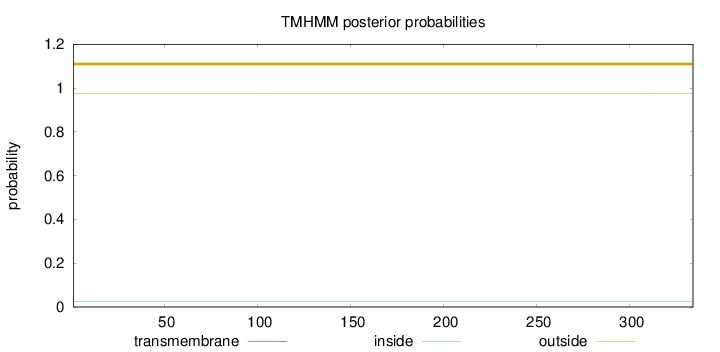
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00135
Exp number, first 60 AAs:
0.000910000000000001
Total prob of N-in:
0.02609
outside
1 - 334
Population Genetic Test Statistics
Pi
306.042
Theta
169.566121
Tajima's D
1.300746
CLR
0.344945
CSRT
0.736013199340033
Interpretation
Uncertain
