Gene
KWMTBOMO04964
Pre Gene Modal
BGIBMGA007990
Annotation
PREDICTED:_protein_ST7_homolog_[Neodiprion_lecontei]
Full name
Suppressor of tumorigenicity 7 protein
Alternative Name
Protein FAM4A1
Protein HELG
Protein HELG
Location in the cell
PlasmaMembrane Reliability : 1.115
Sequence
CDS
ATGTGGGACACAAACGCATATTTGAGCGCATTGACGCCTAAATTCTATGTAGCATTAACAGGCACTTCGTCTTTGATATCAGGACTCATTCTTATTTTCGAATGGTGGTATTTTAGAACATACGGTACCTCATTTATAGAACAAGTATCGCTATCACATCTTGGACCATGGTTAGGTGGCGGCAATGAGACGGATAATTCAAATACTAATACAAACAATACAGCTAGTAATGAATGCAAAGTTTGGCGGAATCCTATGGCTTTACTGCGAGGCGCTGAGTATGGGAGACTTTGGAGTGCCGCAAGGCGAGCTCCACTCACCTATTACGATATGAATTTATCAGCTCAAGACCATCAGGCCTGGTTTAGCTGTGGAGGAGATCAAAGTCCTGTGGAAGAAGCAATGAATCGTGCATGGCGTGAACGTGAACCTACAGCCCGTGTGGCAGCTGCTCGCACTGCACTAGCTAGAGATCCAGACTGTGCTGCTGCATTGTTGTTACTGGCAGAGGAAGATTCACCGACAGTACAAGAGGCTGAACGTGTACTGAAGAGAGCTTGGCGAGCAGCTGAGGCGTCTTGGAGAGCGCACGCAGCCGCTGGATCTGCATATGCTAGCGCAGCCCGACGCGACGCGGCCGTGCTAGCCCACGTCAAGCGTCGAGTTGCTATGTGCGCTCGTCGTCTCGGCAAACTCCGCGACGCCGCTAGAATGTTCCGCGAACTAACGCGAGATGCTCCTCCCGCCCTCCACGCAATGTCACTCCATGAAAACTTGATAGAGGTGCTGTTGGAGCAGAGGCTGTATGCGGACGCGGCTGCTGTTGTTGCGAGATTTGAAGACGCAGGTGCTCCGCCTTCAGCAGCGTTATTGTACACGGCCGCGTTGTTGAGAGCACGTGCTGCCGTCGGCGTTGGTGCACGTCCCGGAGCAGAGGTCATTGCTCTGGAGGCAATACGTCGCGCTATCGAGTTCAATCCTCACGTGCCCGAGTACCTATTAGAATTGCGCGCTTTAACTTTACCACCTGAGCACGTGTTGCGTCGCGGCGACAGCGAAGCGGTCGCGTACGCGTTTTTCCACTTGCGACACTGGCGGAACGCGGAGGGCGCGCTGCGGCTGGTGGCGGCGGCGTGGGCGGGCGCGGAGCGGGCGGGCGCGGGGGCGGGCGCGGGGTGCGCGGCGTGCGCGGACCGCGAGCTGCTTGCGCCGCACCACGCCATGTCCGCATTCCCGCGCCGCCGCGCCGCGCTCCTGCCACTGGCGGCAGCGCTGTGCGCGGGCTGCGCCGTGCTGGCGCTGGCGGCGCACCGGTGGCCGGCGCATGCGCACGCGCTTCTCGACCCGCGCCGCCTGCACTCCCTGTTGGCGGCCGTCTGA
Protein
MWDTNAYLSALTPKFYVALTGTSSLISGLILIFEWWYFRTYGTSFIEQVSLSHLGPWLGGGNETDNSNTNTNNTASNECKVWRNPMALLRGAEYGRLWSAARRAPLTYYDMNLSAQDHQAWFSCGGDQSPVEEAMNRAWREREPTARVAAARTALARDPDCAAALLLLAEEDSPTVQEAERVLKRAWRAAEASWRAHAAAGSAYASAARRDAAVLAHVKRRVAMCARRLGKLRDAARMFRELTRDAPPALHAMSLHENLIEVLLEQRLYADAAAVVARFEDAGAPPSAALLYTAALLRARAAVGVGARPGAEVIALEAIRRAIEFNPHVPEYLLELRALTLPPEHVLRRGDSEAVAYAFFHLRHWRNAEGALRLVAAAWAGAERAGAGAGAGCAACADRELLAPHHAMSAFPRRRAALLPLAAALCAGCAVLALAAHRWPAHAHALLDPRRLHSLLAAV
Summary
Description
May act as a tumor suppressor.
Similarity
Belongs to the ST7 family.
Keywords
Alternative splicing
Complete proteome
Glycoprotein
Membrane
Phosphoprotein
Polymorphism
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Suppressor of tumorigenicity 7 protein
splice variant In isoform 6.
sequence variant In dbSNP:rs201219065.
splice variant In isoform 6.
sequence variant In dbSNP:rs201219065.
Uniprot
H9JEP3
A0A2H1VXJ1
A0A212F0A0
A0A0P6K139
B0X5U6
A0A182H2N3
+ More
A0A0M9A177 A0A0J7P3H8 W5JH00 A0A182FF79 A0A0C9QAR2 E2C7I9 A0A0L7QZS4 A0A3Q1LPG4 E2AM47 A0A1Y1K8J2 A0A146LX50 A0A151WF29 A0A195ATJ9 E9J1K4 F4WKY2 A0A158P3U2 A0A1W4WT20 A0A087ZPH9 A0A2A3EUL2 A0A1Q3F603 A0A336MN84 D6X3H2 A0A154P0B8 A0A026W7Q9 A0A1B0D8Q0 K7J2Z5 A0A232ELU8 A0A1B0CH92 A0A182QKR1 A0A195FJF0 A0A195DZM8 A0A2J7PWR6 A0A067R7W1 A0A182U482 Q7QJH8 A0A182I3E7 A0A182X3M2 A0A182V8I8 A0A182KRF8 A0A182XYZ8 A0A195CNC2 A0A0P4ZEH3 A0A2J7PWR1 A0A182NHR8 A0A164K2P0 A0A182WJ49 A0A0N8A0T2 E9HDV1 A0A0P5TNE7 A0A0P5TK60 A0A2P2I6C2 A0A1B6D6N6 A0A0P6AB91 A0A0P5WUS8 A0A0P5GFE4 A0A1B6M4L4 A0A3R7PEQ7 A0A1B6EYG7 A0A0N7ZJE3 A0A0P4XWZ4 A0A0P5MBD4 A0A0P4W9U9 J3JUX9 A0A1D2MSL3 A0A131Z2E6 L7M774 A0A1E1XDQ3 A0A023GKY1 G3MM84 A0A2R5LKS9 A0A293N6J5 C3YPI6 A0A131XJB9 A0A131Y009 A0A3B3Q998 B7PBY6 A0A0P6I0M7 W5MW97 T1IZ41 W5LB12 A0A3Q3JA04 G3WY21 A0A3B3CDA4 V9KNM4 A0A3B4G615 A0A3P8QKW0 A0A3P9BBA1 A0A2R8ZXV5 Q9NRC1-3 A0A3P8WX59 A0A3Q2VGJ7 A0A3P9HTS5 A0A1A8CUS2 A0A3B4AZN1
A0A0M9A177 A0A0J7P3H8 W5JH00 A0A182FF79 A0A0C9QAR2 E2C7I9 A0A0L7QZS4 A0A3Q1LPG4 E2AM47 A0A1Y1K8J2 A0A146LX50 A0A151WF29 A0A195ATJ9 E9J1K4 F4WKY2 A0A158P3U2 A0A1W4WT20 A0A087ZPH9 A0A2A3EUL2 A0A1Q3F603 A0A336MN84 D6X3H2 A0A154P0B8 A0A026W7Q9 A0A1B0D8Q0 K7J2Z5 A0A232ELU8 A0A1B0CH92 A0A182QKR1 A0A195FJF0 A0A195DZM8 A0A2J7PWR6 A0A067R7W1 A0A182U482 Q7QJH8 A0A182I3E7 A0A182X3M2 A0A182V8I8 A0A182KRF8 A0A182XYZ8 A0A195CNC2 A0A0P4ZEH3 A0A2J7PWR1 A0A182NHR8 A0A164K2P0 A0A182WJ49 A0A0N8A0T2 E9HDV1 A0A0P5TNE7 A0A0P5TK60 A0A2P2I6C2 A0A1B6D6N6 A0A0P6AB91 A0A0P5WUS8 A0A0P5GFE4 A0A1B6M4L4 A0A3R7PEQ7 A0A1B6EYG7 A0A0N7ZJE3 A0A0P4XWZ4 A0A0P5MBD4 A0A0P4W9U9 J3JUX9 A0A1D2MSL3 A0A131Z2E6 L7M774 A0A1E1XDQ3 A0A023GKY1 G3MM84 A0A2R5LKS9 A0A293N6J5 C3YPI6 A0A131XJB9 A0A131Y009 A0A3B3Q998 B7PBY6 A0A0P6I0M7 W5MW97 T1IZ41 W5LB12 A0A3Q3JA04 G3WY21 A0A3B3CDA4 V9KNM4 A0A3B4G615 A0A3P8QKW0 A0A3P9BBA1 A0A2R8ZXV5 Q9NRC1-3 A0A3P8WX59 A0A3Q2VGJ7 A0A3P9HTS5 A0A1A8CUS2 A0A3B4AZN1
Pubmed
19121390
22118469
26999592
26483478
20920257
23761445
+ More
20798317 19393038 28004739 26823975 21282665 21719571 21347285 18362917 19820115 24508170 30249741 20075255 28648823 24845553 12364791 14747013 17210077 20966253 25244985 21292972 22516182 23537049 27289101 26830274 25576852 28503490 22216098 18563158 28049606 29240929 25329095 21709235 29451363 24402279 25186727 22722832 10889047 11279520 14702039 12853948 12690205 15489334 12213198 17974005 16474848 18691976 11726923 24487278 17554307 25463417
20798317 19393038 28004739 26823975 21282665 21719571 21347285 18362917 19820115 24508170 30249741 20075255 28648823 24845553 12364791 14747013 17210077 20966253 25244985 21292972 22516182 23537049 27289101 26830274 25576852 28503490 22216098 18563158 28049606 29240929 25329095 21709235 29451363 24402279 25186727 22722832 10889047 11279520 14702039 12853948 12690205 15489334 12213198 17974005 16474848 18691976 11726923 24487278 17554307 25463417
EMBL
BABH01023622
ODYU01005056
SOQ45559.1
AGBW02011129
OWR47178.1
GDUN01000188
+ More
JAN95731.1 DS232396 EDS41077.1 JXUM01023872 JXUM01048439 KQ561562 KQ560673 KXJ78166.1 KXJ81337.1 KQ435774 KOX75097.1 LBMM01000206 KMR04489.1 ADMH02001422 ETN62598.1 GBYB01011578 JAG81345.1 GL453383 EFN76065.1 KQ414679 KOC64056.1 GL440704 EFN65505.1 GEZM01089028 JAV57759.1 GDHC01006436 JAQ12193.1 KQ983227 KYQ46460.1 KQ976746 KYM75335.1 GL767674 EFZ13204.1 GL888206 EGI65224.1 ADTU01008551 KZ288186 PBC34829.1 GFDL01012055 JAV22990.1 UFQT01000829 SSX27448.1 KQ971374 EEZ97404.1 KQ434787 KZC05282.1 KK107373 QOIP01000004 EZA51661.1 RLU23412.1 AJVK01027636 AJVK01027637 NNAY01003493 OXU19334.1 AJWK01012118 AXCN02000377 KQ981523 KYN40528.1 KQ980044 KYN18079.1 NEVH01020869 PNF20772.1 KK852819 KDR15604.1 AAAB01008807 EAA04741.2 APCN01000207 KQ977513 KYN02213.1 GDIP01213787 JAJ09615.1 PNF20774.1 LRGB01003380 KZS02893.1 GDIP01193681 JAJ29721.1 GL732625 EFX70092.1 GDIP01125188 JAL78526.1 GDIP01125187 JAL78527.1 IACF01003822 LAB69430.1 GEDC01015950 JAS21348.1 GDIP01031950 JAM71765.1 GDIP01081789 JAM21926.1 GDIQ01247540 JAK04185.1 GEBQ01009117 JAT30860.1 QCYY01002638 ROT68745.1 GECZ01026829 GECZ01019905 GECZ01015617 GECZ01004588 JAS42940.1 JAS49864.1 JAS54152.1 JAS65181.1 GDIP01239593 JAI83808.1 GDIP01239592 JAI83809.1 GDIQ01166622 JAK85103.1 GDRN01075207 GDRN01075203 JAI63092.1 BT127044 KB632345 AEE62006.1 ERL93079.1 LJIJ01000593 ODM96023.1 GEDV01003642 JAP84915.1 GACK01004908 JAA60126.1 GFAC01001813 JAT97375.1 GBBM01000877 JAC34541.1 JO842985 AEO34602.1 GGLE01005985 MBY10111.1 GFWV01023312 MAA48039.1 GG666538 EEN57770.1 GEFH01002950 JAP65631.1 GEFM01003958 JAP71838.1 ABJB010842447 ABJB011083945 DS680117 EEC04108.1 GDIQ01011317 JAN83420.1 AHAT01014313 AFFK01020382 AEFK01176138 AEFK01176139 AEFK01176140 JW867318 AFO99835.1 AJFE02025916 AJFE02025917 AJFE02025918 AJFE02025919 AJFE02025920 AF234882 AF234883 AY009152 AY009153 AK093212 CH236947 AC002542 CH471070 BC030954 BC075855 AF415174 CR936656 AJ277291 HADZ01019611 SBP83552.1
JAN95731.1 DS232396 EDS41077.1 JXUM01023872 JXUM01048439 KQ561562 KQ560673 KXJ78166.1 KXJ81337.1 KQ435774 KOX75097.1 LBMM01000206 KMR04489.1 ADMH02001422 ETN62598.1 GBYB01011578 JAG81345.1 GL453383 EFN76065.1 KQ414679 KOC64056.1 GL440704 EFN65505.1 GEZM01089028 JAV57759.1 GDHC01006436 JAQ12193.1 KQ983227 KYQ46460.1 KQ976746 KYM75335.1 GL767674 EFZ13204.1 GL888206 EGI65224.1 ADTU01008551 KZ288186 PBC34829.1 GFDL01012055 JAV22990.1 UFQT01000829 SSX27448.1 KQ971374 EEZ97404.1 KQ434787 KZC05282.1 KK107373 QOIP01000004 EZA51661.1 RLU23412.1 AJVK01027636 AJVK01027637 NNAY01003493 OXU19334.1 AJWK01012118 AXCN02000377 KQ981523 KYN40528.1 KQ980044 KYN18079.1 NEVH01020869 PNF20772.1 KK852819 KDR15604.1 AAAB01008807 EAA04741.2 APCN01000207 KQ977513 KYN02213.1 GDIP01213787 JAJ09615.1 PNF20774.1 LRGB01003380 KZS02893.1 GDIP01193681 JAJ29721.1 GL732625 EFX70092.1 GDIP01125188 JAL78526.1 GDIP01125187 JAL78527.1 IACF01003822 LAB69430.1 GEDC01015950 JAS21348.1 GDIP01031950 JAM71765.1 GDIP01081789 JAM21926.1 GDIQ01247540 JAK04185.1 GEBQ01009117 JAT30860.1 QCYY01002638 ROT68745.1 GECZ01026829 GECZ01019905 GECZ01015617 GECZ01004588 JAS42940.1 JAS49864.1 JAS54152.1 JAS65181.1 GDIP01239593 JAI83808.1 GDIP01239592 JAI83809.1 GDIQ01166622 JAK85103.1 GDRN01075207 GDRN01075203 JAI63092.1 BT127044 KB632345 AEE62006.1 ERL93079.1 LJIJ01000593 ODM96023.1 GEDV01003642 JAP84915.1 GACK01004908 JAA60126.1 GFAC01001813 JAT97375.1 GBBM01000877 JAC34541.1 JO842985 AEO34602.1 GGLE01005985 MBY10111.1 GFWV01023312 MAA48039.1 GG666538 EEN57770.1 GEFH01002950 JAP65631.1 GEFM01003958 JAP71838.1 ABJB010842447 ABJB011083945 DS680117 EEC04108.1 GDIQ01011317 JAN83420.1 AHAT01014313 AFFK01020382 AEFK01176138 AEFK01176139 AEFK01176140 JW867318 AFO99835.1 AJFE02025916 AJFE02025917 AJFE02025918 AJFE02025919 AJFE02025920 AF234882 AF234883 AY009152 AY009153 AK093212 CH236947 AC002542 CH471070 BC030954 BC075855 AF415174 CR936656 AJ277291 HADZ01019611 SBP83552.1
Proteomes
UP000005204
UP000007151
UP000002320
UP000069940
UP000249989
UP000053105
+ More
UP000036403 UP000000673 UP000069272 UP000008237 UP000053825 UP000009136 UP000000311 UP000075809 UP000078540 UP000007755 UP000005205 UP000192223 UP000005203 UP000242457 UP000007266 UP000076502 UP000053097 UP000279307 UP000092462 UP000002358 UP000215335 UP000092461 UP000075886 UP000078541 UP000078492 UP000235965 UP000027135 UP000075902 UP000007062 UP000075840 UP000076407 UP000075903 UP000075882 UP000076408 UP000078542 UP000075884 UP000076858 UP000075920 UP000000305 UP000283509 UP000030742 UP000094527 UP000001554 UP000261540 UP000001555 UP000018468 UP000018467 UP000261600 UP000007648 UP000261560 UP000261460 UP000265100 UP000265160 UP000240080 UP000005640 UP000265120 UP000264840 UP000265200 UP000261520
UP000036403 UP000000673 UP000069272 UP000008237 UP000053825 UP000009136 UP000000311 UP000075809 UP000078540 UP000007755 UP000005205 UP000192223 UP000005203 UP000242457 UP000007266 UP000076502 UP000053097 UP000279307 UP000092462 UP000002358 UP000215335 UP000092461 UP000075886 UP000078541 UP000078492 UP000235965 UP000027135 UP000075902 UP000007062 UP000075840 UP000076407 UP000075903 UP000075882 UP000076408 UP000078542 UP000075884 UP000076858 UP000075920 UP000000305 UP000283509 UP000030742 UP000094527 UP000001554 UP000261540 UP000001555 UP000018468 UP000018467 UP000261600 UP000007648 UP000261560 UP000261460 UP000265100 UP000265160 UP000240080 UP000005640 UP000265120 UP000264840 UP000265200 UP000261520
Pfam
PF04184 ST7
SUPFAM
SSF48452
SSF48452
Gene 3D
CDD
ProteinModelPortal
H9JEP3
A0A2H1VXJ1
A0A212F0A0
A0A0P6K139
B0X5U6
A0A182H2N3
+ More
A0A0M9A177 A0A0J7P3H8 W5JH00 A0A182FF79 A0A0C9QAR2 E2C7I9 A0A0L7QZS4 A0A3Q1LPG4 E2AM47 A0A1Y1K8J2 A0A146LX50 A0A151WF29 A0A195ATJ9 E9J1K4 F4WKY2 A0A158P3U2 A0A1W4WT20 A0A087ZPH9 A0A2A3EUL2 A0A1Q3F603 A0A336MN84 D6X3H2 A0A154P0B8 A0A026W7Q9 A0A1B0D8Q0 K7J2Z5 A0A232ELU8 A0A1B0CH92 A0A182QKR1 A0A195FJF0 A0A195DZM8 A0A2J7PWR6 A0A067R7W1 A0A182U482 Q7QJH8 A0A182I3E7 A0A182X3M2 A0A182V8I8 A0A182KRF8 A0A182XYZ8 A0A195CNC2 A0A0P4ZEH3 A0A2J7PWR1 A0A182NHR8 A0A164K2P0 A0A182WJ49 A0A0N8A0T2 E9HDV1 A0A0P5TNE7 A0A0P5TK60 A0A2P2I6C2 A0A1B6D6N6 A0A0P6AB91 A0A0P5WUS8 A0A0P5GFE4 A0A1B6M4L4 A0A3R7PEQ7 A0A1B6EYG7 A0A0N7ZJE3 A0A0P4XWZ4 A0A0P5MBD4 A0A0P4W9U9 J3JUX9 A0A1D2MSL3 A0A131Z2E6 L7M774 A0A1E1XDQ3 A0A023GKY1 G3MM84 A0A2R5LKS9 A0A293N6J5 C3YPI6 A0A131XJB9 A0A131Y009 A0A3B3Q998 B7PBY6 A0A0P6I0M7 W5MW97 T1IZ41 W5LB12 A0A3Q3JA04 G3WY21 A0A3B3CDA4 V9KNM4 A0A3B4G615 A0A3P8QKW0 A0A3P9BBA1 A0A2R8ZXV5 Q9NRC1-3 A0A3P8WX59 A0A3Q2VGJ7 A0A3P9HTS5 A0A1A8CUS2 A0A3B4AZN1
A0A0M9A177 A0A0J7P3H8 W5JH00 A0A182FF79 A0A0C9QAR2 E2C7I9 A0A0L7QZS4 A0A3Q1LPG4 E2AM47 A0A1Y1K8J2 A0A146LX50 A0A151WF29 A0A195ATJ9 E9J1K4 F4WKY2 A0A158P3U2 A0A1W4WT20 A0A087ZPH9 A0A2A3EUL2 A0A1Q3F603 A0A336MN84 D6X3H2 A0A154P0B8 A0A026W7Q9 A0A1B0D8Q0 K7J2Z5 A0A232ELU8 A0A1B0CH92 A0A182QKR1 A0A195FJF0 A0A195DZM8 A0A2J7PWR6 A0A067R7W1 A0A182U482 Q7QJH8 A0A182I3E7 A0A182X3M2 A0A182V8I8 A0A182KRF8 A0A182XYZ8 A0A195CNC2 A0A0P4ZEH3 A0A2J7PWR1 A0A182NHR8 A0A164K2P0 A0A182WJ49 A0A0N8A0T2 E9HDV1 A0A0P5TNE7 A0A0P5TK60 A0A2P2I6C2 A0A1B6D6N6 A0A0P6AB91 A0A0P5WUS8 A0A0P5GFE4 A0A1B6M4L4 A0A3R7PEQ7 A0A1B6EYG7 A0A0N7ZJE3 A0A0P4XWZ4 A0A0P5MBD4 A0A0P4W9U9 J3JUX9 A0A1D2MSL3 A0A131Z2E6 L7M774 A0A1E1XDQ3 A0A023GKY1 G3MM84 A0A2R5LKS9 A0A293N6J5 C3YPI6 A0A131XJB9 A0A131Y009 A0A3B3Q998 B7PBY6 A0A0P6I0M7 W5MW97 T1IZ41 W5LB12 A0A3Q3JA04 G3WY21 A0A3B3CDA4 V9KNM4 A0A3B4G615 A0A3P8QKW0 A0A3P9BBA1 A0A2R8ZXV5 Q9NRC1-3 A0A3P8WX59 A0A3Q2VGJ7 A0A3P9HTS5 A0A1A8CUS2 A0A3B4AZN1
Ontologies
PANTHER
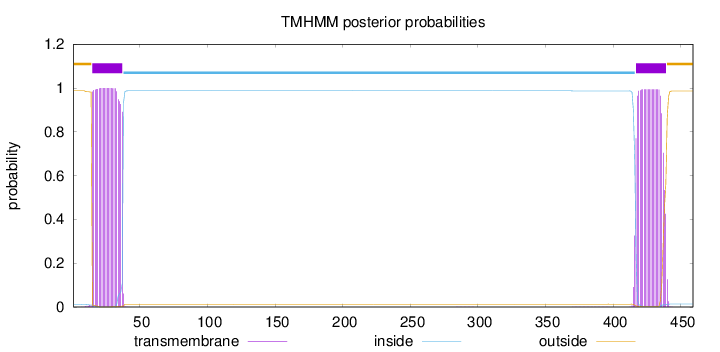
Topology
Subcellular location
Membrane
Length:
459
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.48086
Exp number, first 60 AAs:
22.74213
Total prob of N-in:
0.01249
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 416
TMhelix
417 - 439
outside
440 - 459
Population Genetic Test Statistics
Pi
182.359474
Theta
154.779162
Tajima's D
0.614974
CLR
14.026075
CSRT
0.550822458877056
Interpretation
Uncertain
