Gene
KWMTBOMO04956
Pre Gene Modal
BGIBMGA008082
Annotation
hypothetical_protein_KGM_04336_[Danaus_plexippus]
Full name
Proliferating cell nuclear antigen
Location in the cell
PlasmaMembrane Reliability : 3.798
Sequence
CDS
ATGAAGGCCTCCGATACGTTGCACAGAAACGGTAAAAGCGAGAATGGATCCAAAATTACAGAGAAACACGAACAAAAACATAAAGATCCTTTGACGGAAGGCGTGAAATGGATCGAAAGATACGCCGAAAACTTGGAAAATTTCTTCGAGAGACTACCGGAATTCATTTCCACGTTTATAGCGACTCTGGCGGTGTTTACCTTTGGATCAATTTTAAGAGGAGAGTGGGTCGTCATTCTAGTCACAGCGCTGAAGCAAATCACGGGACACACGCAAACCAAAAATGCCTCAGTCGAGGAAATATTCGAGCTGTTCACATCTAGAAACTTGAAAATGGAGAATTTCAGTATCATATTCATATTAGCTAATGTTGTTTCATTTGGATTTTACTTTATCATCGGAGGATTTTTACATTGGTACTTCTACGTGAAGAGAAGGCACCTCGCCAGTGAATGGAAGATCCAGCCGAACAAATGGCTGTCCCCTGAGTTGGAACGCCACGAAATCATGATAGGAACCCTATCTCTCATTATTACAAGTTCATTCTCAGCTTTTTTAGCCTGTTACATCTTCAATGGGAATCCGTGCACAGTCTATTTTCAGTTCGATGAGTACGGATGGATCTGGTTCTTCTTGCAGTTTCCAGTTATTTTCATTTATATTGATTACACGACCTATATTATGCATCGTCTTTATCACACTCCTTGGTTGTACAAGCATTTCCACAAGCTCCACCATAAGTACAAACAGCCTACAGCCTTTTCTGTAACTGCCATCCATCCAGTAGAAATTATGCATATCCAACTAACGATGTGTCTACCTCTCTTCACAATACCTACACATTGGGCATCATTCTACGCTGTGGCCATTTACAATTACTACCACGGCATCATTGATCATTCCGGCATCAATTTTAAGGCCCAGTGGTGGCAGCCGTGGCAGCCTGACGCCGAATTCCATGATCAGCATCACGAATTTTTCCATTGCAACTTTGGCTTCAATATGTATCTGTGGGACAAGTGGCACGGTACGATGAGGAAGCAGTACAACGTGTACACCGAAGAGACATTCCACGGTGAGGCTCCGTCTGCAGAAACGGCCGAAGGAAAGGCGATATTGGAAGCGAACCCCGAACTTATTGATAGTATCAACAAACCATTACTGACAAAGACTGAAACCCAAAAAACTAAATGA
Protein
MKASDTLHRNGKSENGSKITEKHEQKHKDPLTEGVKWIERYAENLENFFERLPEFISTFIATLAVFTFGSILRGEWVVILVTALKQITGHTQTKNASVEEIFELFTSRNLKMENFSIIFILANVVSFGFYFIIGGFLHWYFYVKRRHLASEWKIQPNKWLSPELERHEIMIGTLSLIITSSFSAFLACYIFNGNPCTVYFQFDEYGWIWFFLQFPVIFIYIDYTTYIMHRLYHTPWLYKHFHKLHHKYKQPTAFSVTAIHPVEIMHIQLTMCLPLFTIPTHWASFYAVAIYNYYHGIIDHSGINFKAQWWQPWQPDAEFHDQHHEFFHCNFGFNMYLWDKWHGTMRKQYNVYTEETFHGEAPSAETAEGKAILEANPELIDSINKPLLTKTETQKTK
Summary
Description
This protein is an auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand.
Similarity
Belongs to the sterol desaturase family.
Belongs to the PCNA family.
Belongs to the PCNA family.
Uniprot
H9JEY5
A0A212EYU2
A0A194QD32
A0A2H1X1S0
A0A3S2L4W3
A0A2A4JHB7
+ More
A0A194QQX8 A0A0L7LLQ6 D6W8U5 A0A067REM4 A0A1B0D5I1 A0A0T6BHB5 A0A1B6M8U1 V5GYV5 A0A2J7QVD1 J3JUC1 A0A1B0CE06 U4UN31 A0A2P8Y2F5 A0A0P5TR20 A0A0P5TRA2 A0A162NV58 A0A0P5FSY0 A0A0P5E638 A0A0P5CA23 A0A0P5BHW8 A0A0P6H9R4 A0A232EQP5 A0A1D2M1S0 A0A226EP73 E9G489 A0A1W4WN39 A0A1B6HK04 A0A0P6D719 A0A0P6IWD3 A0A0P6BFG3 A0A3R7Q9Z7 A0A0P6FW02 A0A0P5LKG2 A0A0N8D6M9 A0A0P6GFD7 A0A0P5JWC5 A0A0P5KAY6 A0A0N8BIG8 A0A0N8CCW4 A0A0P5P9Z7 A0A0P5YA60 A0A0P5WUF1 A0A0P5NVJ5 A0A0P5S6L9 A0A0N8EEW7 A0A1S3D674 A0A1B6DA27 A0A0P5ACZ6 A0A1S3D6F5 A0A0N8CZZ3 A0A0P5LYT1 A0A1S3JHH3 A0A1S3IIK0 A0A0P5M3B3 A0A0P5U9P3 A7RXC0 A0A2H8TSA6 A0A2S2PA06 A0A087TGT2 A0A087U4C2 J9JVH2 A0A0N8CKJ3 A0A2S2PX93 A0A210Q444 R7UVA9 C3XWE8 F6TVN3 R7TEA4 W4YN73 W4YCZ2
A0A194QQX8 A0A0L7LLQ6 D6W8U5 A0A067REM4 A0A1B0D5I1 A0A0T6BHB5 A0A1B6M8U1 V5GYV5 A0A2J7QVD1 J3JUC1 A0A1B0CE06 U4UN31 A0A2P8Y2F5 A0A0P5TR20 A0A0P5TRA2 A0A162NV58 A0A0P5FSY0 A0A0P5E638 A0A0P5CA23 A0A0P5BHW8 A0A0P6H9R4 A0A232EQP5 A0A1D2M1S0 A0A226EP73 E9G489 A0A1W4WN39 A0A1B6HK04 A0A0P6D719 A0A0P6IWD3 A0A0P6BFG3 A0A3R7Q9Z7 A0A0P6FW02 A0A0P5LKG2 A0A0N8D6M9 A0A0P6GFD7 A0A0P5JWC5 A0A0P5KAY6 A0A0N8BIG8 A0A0N8CCW4 A0A0P5P9Z7 A0A0P5YA60 A0A0P5WUF1 A0A0P5NVJ5 A0A0P5S6L9 A0A0N8EEW7 A0A1S3D674 A0A1B6DA27 A0A0P5ACZ6 A0A1S3D6F5 A0A0N8CZZ3 A0A0P5LYT1 A0A1S3JHH3 A0A1S3IIK0 A0A0P5M3B3 A0A0P5U9P3 A7RXC0 A0A2H8TSA6 A0A2S2PA06 A0A087TGT2 A0A087U4C2 J9JVH2 A0A0N8CKJ3 A0A2S2PX93 A0A210Q444 R7UVA9 C3XWE8 F6TVN3 R7TEA4 W4YN73 W4YCZ2
Pubmed
EMBL
BABH01023556
BABH01023557
BABH01023558
BABH01023559
AGBW02011440
OWR46668.1
+ More
KQ459185 KPJ03329.1 ODYU01012752 SOQ59217.1 RSAL01000150 RVE45782.1 NWSH01001575 PCG70823.1 KQ461198 KPJ05936.1 JTDY01000669 KOB76279.1 KQ971312 EEZ98257.2 KK852746 KDR17319.1 AJVK01025282 LJIG01000201 KRT86739.1 GEBQ01007713 JAT32264.1 GALX01002833 JAB65633.1 NEVH01010477 PNF32539.1 BT126834 AEE61796.1 AJWK01008460 KB632375 ERL93863.1 PYGN01001017 PSN38445.1 GDIP01122754 JAL80960.1 GDIP01122755 JAL80959.1 LRGB01000531 KZS18293.1 GDIP01144151 JAJ79251.1 GDIP01147554 JAJ75848.1 GDIP01189692 JAJ33710.1 GDIP01189691 JAJ33711.1 GDIQ01044327 JAN50410.1 NNAY01002726 OXU20684.1 LJIJ01006772 ODM86918.1 LNIX01000002 OXA59435.1 GL732531 EFX86052.1 GECU01032698 JAS75008.1 GDIQ01081854 JAN12883.1 GDIQ01003587 JAN91150.1 GDIP01030061 JAM73654.1 QCYY01002156 ROT72445.1 GDIQ01042018 JAN52719.1 GDIQ01170583 JAK81142.1 GDIP01063615 JAM40100.1 GDIQ01044328 JAN50409.1 GDIQ01194260 JAK57465.1 GDIQ01212743 JAK38982.1 GDIQ01174515 JAK77210.1 GDIQ01092147 JAL59579.1 GDIQ01153177 JAK98548.1 GDIP01060659 JAM43056.1 GDIP01093549 JAM10166.1 GDIQ01137097 JAL14629.1 GDIQ01092148 JAL59578.1 GDIQ01033532 JAN61205.1 GEDC01014751 JAS22547.1 GDIP01200743 JAJ22659.1 GDIP01082303 JAM21412.1 GDIQ01172226 JAK79499.1 GDIQ01161372 JAK90353.1 GDIP01117739 JAL85975.1 DS469549 EDO44003.1 GFXV01005044 MBW16849.1 GGMR01013672 MBY26291.1 KK115165 KFM64321.1 KK118087 KFM72211.1 ABLF02027362 GDIP01122703 JAL81011.1 GGMS01000945 MBY70148.1 NEDP02005089 OWF43495.1 AMQN01006015 KB297391 ELU10573.1 GG666471 EEN67682.1 AMQN01013603 KB310334 ELT91807.1 AAGJ04030492 AAGJ04030495
KQ459185 KPJ03329.1 ODYU01012752 SOQ59217.1 RSAL01000150 RVE45782.1 NWSH01001575 PCG70823.1 KQ461198 KPJ05936.1 JTDY01000669 KOB76279.1 KQ971312 EEZ98257.2 KK852746 KDR17319.1 AJVK01025282 LJIG01000201 KRT86739.1 GEBQ01007713 JAT32264.1 GALX01002833 JAB65633.1 NEVH01010477 PNF32539.1 BT126834 AEE61796.1 AJWK01008460 KB632375 ERL93863.1 PYGN01001017 PSN38445.1 GDIP01122754 JAL80960.1 GDIP01122755 JAL80959.1 LRGB01000531 KZS18293.1 GDIP01144151 JAJ79251.1 GDIP01147554 JAJ75848.1 GDIP01189692 JAJ33710.1 GDIP01189691 JAJ33711.1 GDIQ01044327 JAN50410.1 NNAY01002726 OXU20684.1 LJIJ01006772 ODM86918.1 LNIX01000002 OXA59435.1 GL732531 EFX86052.1 GECU01032698 JAS75008.1 GDIQ01081854 JAN12883.1 GDIQ01003587 JAN91150.1 GDIP01030061 JAM73654.1 QCYY01002156 ROT72445.1 GDIQ01042018 JAN52719.1 GDIQ01170583 JAK81142.1 GDIP01063615 JAM40100.1 GDIQ01044328 JAN50409.1 GDIQ01194260 JAK57465.1 GDIQ01212743 JAK38982.1 GDIQ01174515 JAK77210.1 GDIQ01092147 JAL59579.1 GDIQ01153177 JAK98548.1 GDIP01060659 JAM43056.1 GDIP01093549 JAM10166.1 GDIQ01137097 JAL14629.1 GDIQ01092148 JAL59578.1 GDIQ01033532 JAN61205.1 GEDC01014751 JAS22547.1 GDIP01200743 JAJ22659.1 GDIP01082303 JAM21412.1 GDIQ01172226 JAK79499.1 GDIQ01161372 JAK90353.1 GDIP01117739 JAL85975.1 DS469549 EDO44003.1 GFXV01005044 MBW16849.1 GGMR01013672 MBY26291.1 KK115165 KFM64321.1 KK118087 KFM72211.1 ABLF02027362 GDIP01122703 JAL81011.1 GGMS01000945 MBY70148.1 NEDP02005089 OWF43495.1 AMQN01006015 KB297391 ELU10573.1 GG666471 EEN67682.1 AMQN01013603 KB310334 ELT91807.1 AAGJ04030492 AAGJ04030495
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000218220
UP000053240
+ More
UP000037510 UP000007266 UP000027135 UP000092462 UP000235965 UP000092461 UP000030742 UP000245037 UP000076858 UP000215335 UP000094527 UP000198287 UP000000305 UP000192223 UP000283509 UP000079169 UP000085678 UP000001593 UP000054359 UP000007819 UP000242188 UP000014760 UP000001554 UP000008144 UP000007110
UP000037510 UP000007266 UP000027135 UP000092462 UP000235965 UP000092461 UP000030742 UP000245037 UP000076858 UP000215335 UP000094527 UP000198287 UP000000305 UP000192223 UP000283509 UP000079169 UP000085678 UP000001593 UP000054359 UP000007819 UP000242188 UP000014760 UP000001554 UP000008144 UP000007110
Interpro
ProteinModelPortal
H9JEY5
A0A212EYU2
A0A194QD32
A0A2H1X1S0
A0A3S2L4W3
A0A2A4JHB7
+ More
A0A194QQX8 A0A0L7LLQ6 D6W8U5 A0A067REM4 A0A1B0D5I1 A0A0T6BHB5 A0A1B6M8U1 V5GYV5 A0A2J7QVD1 J3JUC1 A0A1B0CE06 U4UN31 A0A2P8Y2F5 A0A0P5TR20 A0A0P5TRA2 A0A162NV58 A0A0P5FSY0 A0A0P5E638 A0A0P5CA23 A0A0P5BHW8 A0A0P6H9R4 A0A232EQP5 A0A1D2M1S0 A0A226EP73 E9G489 A0A1W4WN39 A0A1B6HK04 A0A0P6D719 A0A0P6IWD3 A0A0P6BFG3 A0A3R7Q9Z7 A0A0P6FW02 A0A0P5LKG2 A0A0N8D6M9 A0A0P6GFD7 A0A0P5JWC5 A0A0P5KAY6 A0A0N8BIG8 A0A0N8CCW4 A0A0P5P9Z7 A0A0P5YA60 A0A0P5WUF1 A0A0P5NVJ5 A0A0P5S6L9 A0A0N8EEW7 A0A1S3D674 A0A1B6DA27 A0A0P5ACZ6 A0A1S3D6F5 A0A0N8CZZ3 A0A0P5LYT1 A0A1S3JHH3 A0A1S3IIK0 A0A0P5M3B3 A0A0P5U9P3 A7RXC0 A0A2H8TSA6 A0A2S2PA06 A0A087TGT2 A0A087U4C2 J9JVH2 A0A0N8CKJ3 A0A2S2PX93 A0A210Q444 R7UVA9 C3XWE8 F6TVN3 R7TEA4 W4YN73 W4YCZ2
A0A194QQX8 A0A0L7LLQ6 D6W8U5 A0A067REM4 A0A1B0D5I1 A0A0T6BHB5 A0A1B6M8U1 V5GYV5 A0A2J7QVD1 J3JUC1 A0A1B0CE06 U4UN31 A0A2P8Y2F5 A0A0P5TR20 A0A0P5TRA2 A0A162NV58 A0A0P5FSY0 A0A0P5E638 A0A0P5CA23 A0A0P5BHW8 A0A0P6H9R4 A0A232EQP5 A0A1D2M1S0 A0A226EP73 E9G489 A0A1W4WN39 A0A1B6HK04 A0A0P6D719 A0A0P6IWD3 A0A0P6BFG3 A0A3R7Q9Z7 A0A0P6FW02 A0A0P5LKG2 A0A0N8D6M9 A0A0P6GFD7 A0A0P5JWC5 A0A0P5KAY6 A0A0N8BIG8 A0A0N8CCW4 A0A0P5P9Z7 A0A0P5YA60 A0A0P5WUF1 A0A0P5NVJ5 A0A0P5S6L9 A0A0N8EEW7 A0A1S3D674 A0A1B6DA27 A0A0P5ACZ6 A0A1S3D6F5 A0A0N8CZZ3 A0A0P5LYT1 A0A1S3JHH3 A0A1S3IIK0 A0A0P5M3B3 A0A0P5U9P3 A7RXC0 A0A2H8TSA6 A0A2S2PA06 A0A087TGT2 A0A087U4C2 J9JVH2 A0A0N8CKJ3 A0A2S2PX93 A0A210Q444 R7UVA9 C3XWE8 F6TVN3 R7TEA4 W4YN73 W4YCZ2
Ontologies
GO
Topology
Subcellular location
Nucleus
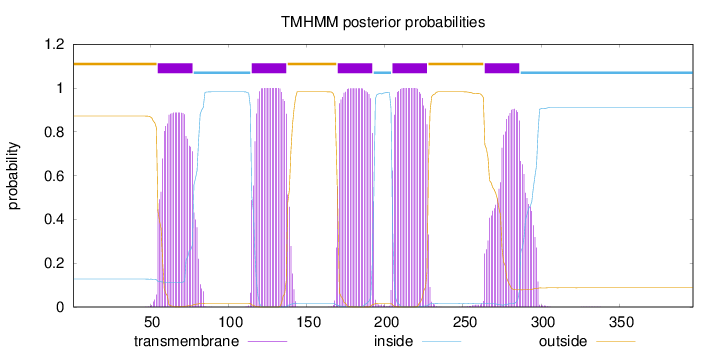
Length:
397
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
108.67854
Exp number, first 60 AAs:
3.90205
Total prob of N-in:
0.12783
outside
1 - 54
TMhelix
55 - 77
inside
78 - 114
TMhelix
115 - 137
outside
138 - 169
TMhelix
170 - 192
inside
193 - 204
TMhelix
205 - 227
outside
228 - 263
TMhelix
264 - 286
inside
287 - 397
Population Genetic Test Statistics
Pi
239.374439
Theta
189.077479
Tajima's D
0.491771
CLR
0.613333
CSRT
0.507724613769312
Interpretation
Uncertain
