Gene
KWMTBOMO04954
Pre Gene Modal
BGIBMGA008080
Annotation
putative_zinc_finger_protein_nocA_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 3.778
Sequence
CDS
ATGGTGGTGCTTGAAGACGGTGGAATGATGGCTACAAATCCCAACCAGTACTTACAACCAGATTATTTAGCACCGCTTCCGTCCACTTTGGACTCGAAAAAAAGCCCGCTTGCGCTTCTAGCGCAAACCTGCAGCCAAATTGGTGTTGACTCGACACCTTCAAAGCCGCTGCTCCCACCACACGACAAGAAGAAAGTGAACAGTGTTAATAACAGTGATATAATAAGTCGTTCGTCACCAAACAGTAATACAATGAAACAAGACAAGCCGCGATCGTCTCCGGAAAGCAAACATTTGGCGTTCAAGCCATACGAGGCAAACGTTATAACAAAAAAACCGGATGAGACTCGGCCTTCTTCCAAAGCTAGTTCGGACAGTTCCGAAGACAAGAAATCTGGAAAGAGCACCCCGGGACGTAAATCAACATCGCCCAGTGGTGAAAATGGAAAAAACAGTCCTTCAAATGAAACAAAATCTTCTTCAGCCAGTTCATCTGGAACAAGCCCTATAATCCGCTCCGGATTAGAGGTTTTGGGACACGGCAAGGATCATCTGGGCGCTTTCAAGAATATACCTGGATTACCAGGATTTAATCCACTCTCTGGACTTTGTTGCCCGCCTGGATTGGAGCAGCACGGAAACCCGGCATTCAGACCGCCGTATGCCGGAGCACCATTCAATGCTCATCATGCCGCTATGCTTGCCGCCGCAGCTGCTTTTCCCGGATCTTCAGCAAACCCATACCTTGGATATGCGAGAGTTAAAACACCTGCCGGGGGAGAGACGTTGGTCCCTGTCTGCAAAGATCCATATTGCACAGGCTGTCAGTTCTCAGTCAACAACCATCATCTGTTAATGAGCAATGGATCTTGCCCCGCCGGGTGCACACAATGCGAACATCAAAAGTACAACTTAGCAATGGCAATGGCTCTGTCACAGCAAGGAGCCGCCGCTAGTAGTTTGCCGTACCACCAGCTGAGCCGACCCTACGTTTGCAACTGGATCGTCGGCGAATCTTACTGCGGCAAGCGGTTTGGAAATTCCGAGGAATTGTTACAGCACTTGAGAAGCCACACAGCCGACGGATCCGCTCCATCGACCAGTGCATCGACTCAAGCGTCCATTTTAAATCAATTTAATCCACTGTTCACGACTGCCGGTCTTCGTAGCGCATACCCGACGGCGCCGCTCAGCCCACTTTCTGCGAGTAGATACCATCCTTACTCAAAAGCTGGTCTCCCGGCCAGCCTCAGTGCTTCACCATACGGTGCCTTCAACCCTGCGCTCGGACCGTTTTATTCTCCCTACGCCATGTATGGACAGAGACTTGGGGCGGCAGCCGTTCACCAGTAA
Protein
MVVLEDGGMMATNPNQYLQPDYLAPLPSTLDSKKSPLALLAQTCSQIGVDSTPSKPLLPPHDKKKVNSVNNSDIISRSSPNSNTMKQDKPRSSPESKHLAFKPYEANVITKKPDETRPSSKASSDSSEDKKSGKSTPGRKSTSPSGENGKNSPSNETKSSSASSSGTSPIIRSGLEVLGHGKDHLGAFKNIPGLPGFNPLSGLCCPPGLEQHGNPAFRPPYAGAPFNAHHAAMLAAAAAFPGSSANPYLGYARVKTPAGGETLVPVCKDPYCTGCQFSVNNHHLLMSNGSCPAGCTQCEHQKYNLAMAMALSQQGAAASSLPYHQLSRPYVCNWIVGESYCGKRFGNSEELLQHLRSHTADGSAPSTSASTQASILNQFNPLFTTAGLRSAYPTAPLSPLSASRYHPYSKAGLPASLSASPYGAFNPALGPFYSPYAMYGQRLGAAAVHQ
Summary
Uniprot
H9JEY3
A0A3S2NQC8
A0A2A4JGV7
A0A2H1WKD6
A0A2W1BIT0
A0A194QK75
+ More
A0A212EYV0 A0A0L7LFK6 A0A194QEF6 D6W8V3 A0A1W4WN69 A0A1Y1KWX1 A0A1B0DBX5 A0A1L8DD42 A0A0T6B9X9 A0A1B6BX88 A0A0C9R8B6 E0VDX6 A0A0L7R3G4 F4WAL4 A0A0J7L520 A0A1B6MCU0 A0A2J7QVD6 A0A0V0GA86 A0A224XQF8 A0A067R7U4 A0A023EY87 A0A0P4VSQ9 T1HG55 A0A0A9VXX9 B0W165 Q1DGT6 A0A088A7M8 A0A0M8ZZR6 A0A2P8Y3V3 A0A1L8DBL9 A0A1W4UJ93 N6UPX2 U4UDM3 A0A1B6G6U1 E2AK20 A0A2R7WHP3 A0A2P2HYT4 A0A1S3DRT4 A0A182FMV6 A0A2M4BI66 A0A2M3YZW4
A0A212EYV0 A0A0L7LFK6 A0A194QEF6 D6W8V3 A0A1W4WN69 A0A1Y1KWX1 A0A1B0DBX5 A0A1L8DD42 A0A0T6B9X9 A0A1B6BX88 A0A0C9R8B6 E0VDX6 A0A0L7R3G4 F4WAL4 A0A0J7L520 A0A1B6MCU0 A0A2J7QVD6 A0A0V0GA86 A0A224XQF8 A0A067R7U4 A0A023EY87 A0A0P4VSQ9 T1HG55 A0A0A9VXX9 B0W165 Q1DGT6 A0A088A7M8 A0A0M8ZZR6 A0A2P8Y3V3 A0A1L8DBL9 A0A1W4UJ93 N6UPX2 U4UDM3 A0A1B6G6U1 E2AK20 A0A2R7WHP3 A0A2P2HYT4 A0A1S3DRT4 A0A182FMV6 A0A2M4BI66 A0A2M3YZW4
Pubmed
EMBL
BABH01023540
BABH01023541
RSAL01000150
RVE45781.1
NWSH01001575
PCG70824.1
+ More
ODYU01009256 SOQ53541.1 KZ150029 PZC74768.1 KQ461198 KPJ05937.1 AGBW02011440 OWR46666.1 JTDY01001336 KOB74160.1 KQ459185 KPJ03330.1 KQ971312 EEZ99225.1 GEZM01077219 JAV63367.1 AJVK01013837 GFDF01009705 JAV04379.1 LJIG01002787 KRT84153.1 GEDC01031412 JAS05886.1 GBYB01003056 JAG72823.1 DS235088 EEB11582.1 KQ414663 KOC65417.1 GL888050 EGI68772.1 LBMM01000746 KMQ97584.1 GEBQ01006226 JAT33751.1 NEVH01010477 PNF32544.1 GECL01001049 JAP05075.1 GFTR01005624 JAW10802.1 KK852642 KDR19597.1 GBBI01004675 JAC14037.1 GDKW01001080 JAI55515.1 ACPB03012233 GBHO01043578 GBRD01009559 GDHC01018503 JAG00026.1 JAG56265.1 JAQ00126.1 DS231820 EDS44039.1 CH900432 CH477392 EAT32366.1 EAT41939.1 KQ435789 KOX74430.1 PYGN01000965 PSN38864.1 GFDF01010226 JAV03858.1 APGK01021796 KB740364 ENN80787.1 KB632275 ERL91157.1 GECZ01011611 JAS58158.1 GL440135 EFN66230.1 KK854842 PTY19162.1 IACF01001221 LAB66935.1 GGFJ01003542 MBW52683.1 GGFM01001056 MBW21807.1
ODYU01009256 SOQ53541.1 KZ150029 PZC74768.1 KQ461198 KPJ05937.1 AGBW02011440 OWR46666.1 JTDY01001336 KOB74160.1 KQ459185 KPJ03330.1 KQ971312 EEZ99225.1 GEZM01077219 JAV63367.1 AJVK01013837 GFDF01009705 JAV04379.1 LJIG01002787 KRT84153.1 GEDC01031412 JAS05886.1 GBYB01003056 JAG72823.1 DS235088 EEB11582.1 KQ414663 KOC65417.1 GL888050 EGI68772.1 LBMM01000746 KMQ97584.1 GEBQ01006226 JAT33751.1 NEVH01010477 PNF32544.1 GECL01001049 JAP05075.1 GFTR01005624 JAW10802.1 KK852642 KDR19597.1 GBBI01004675 JAC14037.1 GDKW01001080 JAI55515.1 ACPB03012233 GBHO01043578 GBRD01009559 GDHC01018503 JAG00026.1 JAG56265.1 JAQ00126.1 DS231820 EDS44039.1 CH900432 CH477392 EAT32366.1 EAT41939.1 KQ435789 KOX74430.1 PYGN01000965 PSN38864.1 GFDF01010226 JAV03858.1 APGK01021796 KB740364 ENN80787.1 KB632275 ERL91157.1 GECZ01011611 JAS58158.1 GL440135 EFN66230.1 KK854842 PTY19162.1 IACF01001221 LAB66935.1 GGFJ01003542 MBW52683.1 GGFM01001056 MBW21807.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000053268 UP000007266 UP000192223 UP000092462 UP000009046 UP000053825 UP000007755 UP000036403 UP000235965 UP000027135 UP000015103 UP000002320 UP000008820 UP000005203 UP000053105 UP000245037 UP000192221 UP000019118 UP000030742 UP000000311 UP000079169 UP000069272
UP000053268 UP000007266 UP000192223 UP000092462 UP000009046 UP000053825 UP000007755 UP000036403 UP000235965 UP000027135 UP000015103 UP000002320 UP000008820 UP000005203 UP000053105 UP000245037 UP000192221 UP000019118 UP000030742 UP000000311 UP000079169 UP000069272
PRIDE
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JEY3
A0A3S2NQC8
A0A2A4JGV7
A0A2H1WKD6
A0A2W1BIT0
A0A194QK75
+ More
A0A212EYV0 A0A0L7LFK6 A0A194QEF6 D6W8V3 A0A1W4WN69 A0A1Y1KWX1 A0A1B0DBX5 A0A1L8DD42 A0A0T6B9X9 A0A1B6BX88 A0A0C9R8B6 E0VDX6 A0A0L7R3G4 F4WAL4 A0A0J7L520 A0A1B6MCU0 A0A2J7QVD6 A0A0V0GA86 A0A224XQF8 A0A067R7U4 A0A023EY87 A0A0P4VSQ9 T1HG55 A0A0A9VXX9 B0W165 Q1DGT6 A0A088A7M8 A0A0M8ZZR6 A0A2P8Y3V3 A0A1L8DBL9 A0A1W4UJ93 N6UPX2 U4UDM3 A0A1B6G6U1 E2AK20 A0A2R7WHP3 A0A2P2HYT4 A0A1S3DRT4 A0A182FMV6 A0A2M4BI66 A0A2M3YZW4
A0A212EYV0 A0A0L7LFK6 A0A194QEF6 D6W8V3 A0A1W4WN69 A0A1Y1KWX1 A0A1B0DBX5 A0A1L8DD42 A0A0T6B9X9 A0A1B6BX88 A0A0C9R8B6 E0VDX6 A0A0L7R3G4 F4WAL4 A0A0J7L520 A0A1B6MCU0 A0A2J7QVD6 A0A0V0GA86 A0A224XQF8 A0A067R7U4 A0A023EY87 A0A0P4VSQ9 T1HG55 A0A0A9VXX9 B0W165 Q1DGT6 A0A088A7M8 A0A0M8ZZR6 A0A2P8Y3V3 A0A1L8DBL9 A0A1W4UJ93 N6UPX2 U4UDM3 A0A1B6G6U1 E2AK20 A0A2R7WHP3 A0A2P2HYT4 A0A1S3DRT4 A0A182FMV6 A0A2M4BI66 A0A2M3YZW4
PDB
1VA2
E-value=0.000429967,
Score=104
Ontologies
GO
Topology
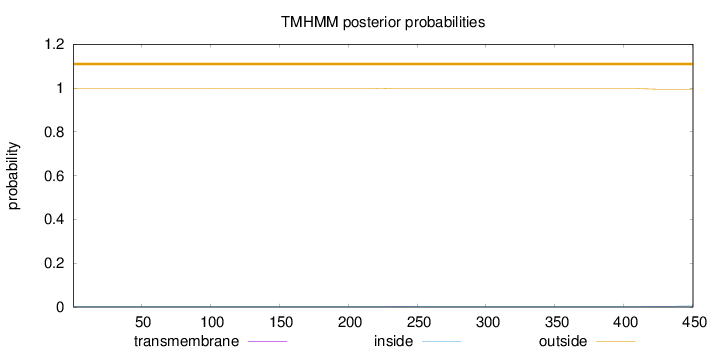
Length:
450
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08764
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00129
outside
1 - 450
Population Genetic Test Statistics
Pi
227.782321
Theta
189.325029
Tajima's D
0.727108
CLR
0
CSRT
0.585070746462677
Interpretation
Uncertain
