Gene
KWMTBOMO04951
Pre Gene Modal
BGIBMGA007996
Annotation
PREDICTED:_pickpocket_protein_28-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.16 Mitochondrial Reliability : 1.435 Nuclear Reliability : 1.63
Sequence
CDS
ATGTTTTTTTTTAGTTTACATGGAGATTTTGCTTACTTGGAATCAACAGAGATGAGCCGTAATTGGACGATAGAGGAAGGCTATAAAAGCGAAGATAACAATACATATCCAAGAAGAGGAAGGAAAAACGGGGCCGAGCCAGATTTAAAAGTTGTACTTAAAGATAACCCGGAACAGAAAGATAAGCTATGTAATAAACTTAAAAATGGTTTCAAGATATTTATACACCATCCCGCAGATTTGCCGCAGCCTTCATTGTACTATTACAGTGCGTTGTCGTCCCAAGTTTCTTCTATGGCACTAAAATTTGATATTTTGCGTACATCAAACAGTTTGGATTCATTACCCGTTGAGCAGTAA
Protein
MFFFSLHGDFAYLESTEMSRNWTIEEGYKSEDNNTYPRRGRKNGAEPDLKVVLKDNPEQKDKLCNKLKNGFKIFIHHPADLPQPSLYYYSALSSQVSSMALKFDILRTSNSLDSLPVEQ
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9JEP9
A0A0L7LLW2
A0A0L7L972
A0A3S2LWT2
A0A0L7L821
A0A0L7KTM8
+ More
A0A0L7KMU4 A0A2H1X321 A0A2A4JK97 A0A2W1BBD0 A0A2W1BGB8 D6WXM6 A0A194R8M9 H9JHM1 A0A2H1WS18 A0A194PW83 A0A3S2NPU4 A0A1J1HMY9 A0A212EHF3 B0WKH8 A0A1B0DLX3 B0WKI1 A0A1B0DLX5 B0WDC3 A0A1Y1N7B1 B0WKI4 A0A1Y1N7X2 A0A1Y1NA93 A0A182H7C8 A0A1Y1N7C2 Q16GX7 A0A1Y1NB01 A0A182J6D0 A0A1Y1N7G4 A0A1S4G1A5 A0A1W4XR61 A0A182W972 N6U8E9 A0A182QS19 U4UGG3 U4UN62 N6TUG2 A0A182S9T8 B4ML83 Q7Q066 A0A182MLZ3 B0X0R9 A0A182R5F2 A0A182THH2 A0A182Y1U0 A0A182HQ19 A0A182XD77 A0A182UXK6 A0A182KSP4 A0A182K171 T1H5I3 A0A182IW77 A0A1B0GL64 A0A182WEP0 A0A182FU38 A0A182MYI9 A0A1I8M8B2 A0A1I8PG23 W5JNH9 A0A084WHX7 A0A139WCP8 A0A139WKH1 A0A1W4U6V2 B4KYR2 Q17DP0 A0A1I8Q829 A0A084WGS5 A0A0L0CPJ6 A0A1S4F6X7 Q9VS73 B3NFC4 B4HV75 A0A0J9RQW9 B0X0S0 A0A182JZW4 A0A182YLS3 A0A3B0JWU3 B3M5Z2 A0A1B0GH50 A0A139WCM4 A0A084W8C3 A0A182GCI9 B4PC79 A0A182FW18 A0A1I8NHK4 D2A5Z0 A0A182MGT5 A0A182PBQ5 A0A182KHA8 A0A182N248
A0A0L7KMU4 A0A2H1X321 A0A2A4JK97 A0A2W1BBD0 A0A2W1BGB8 D6WXM6 A0A194R8M9 H9JHM1 A0A2H1WS18 A0A194PW83 A0A3S2NPU4 A0A1J1HMY9 A0A212EHF3 B0WKH8 A0A1B0DLX3 B0WKI1 A0A1B0DLX5 B0WDC3 A0A1Y1N7B1 B0WKI4 A0A1Y1N7X2 A0A1Y1NA93 A0A182H7C8 A0A1Y1N7C2 Q16GX7 A0A1Y1NB01 A0A182J6D0 A0A1Y1N7G4 A0A1S4G1A5 A0A1W4XR61 A0A182W972 N6U8E9 A0A182QS19 U4UGG3 U4UN62 N6TUG2 A0A182S9T8 B4ML83 Q7Q066 A0A182MLZ3 B0X0R9 A0A182R5F2 A0A182THH2 A0A182Y1U0 A0A182HQ19 A0A182XD77 A0A182UXK6 A0A182KSP4 A0A182K171 T1H5I3 A0A182IW77 A0A1B0GL64 A0A182WEP0 A0A182FU38 A0A182MYI9 A0A1I8M8B2 A0A1I8PG23 W5JNH9 A0A084WHX7 A0A139WCP8 A0A139WKH1 A0A1W4U6V2 B4KYR2 Q17DP0 A0A1I8Q829 A0A084WGS5 A0A0L0CPJ6 A0A1S4F6X7 Q9VS73 B3NFC4 B4HV75 A0A0J9RQW9 B0X0S0 A0A182JZW4 A0A182YLS3 A0A3B0JWU3 B3M5Z2 A0A1B0GH50 A0A139WCM4 A0A084W8C3 A0A182GCI9 B4PC79 A0A182FW18 A0A1I8NHK4 D2A5Z0 A0A182MGT5 A0A182PBQ5 A0A182KHA8 A0A182N248
Pubmed
EMBL
BABH01023503
BABH01023504
BABH01023505
JTDY01000591
KOB76548.1
JTDY01002194
+ More
KOB71904.1 RSAL01000150 RVE45780.1 JTDY01002335 KOB71642.1 JTDY01005791 KOB66623.1 JTDY01008958 KOB64264.1 ODYU01012618 SOQ59034.1 NWSH01001201 PCG72148.1 KZ150309 PZC71475.1 KZ150082 PZC73838.1 KQ971362 EFA08873.2 KQ460597 KPJ13859.1 BABH01012830 ODYU01010562 SOQ55762.1 KQ459591 KPI97019.1 RSAL01000014 RVE53205.1 CVRI01000004 CRK87601.1 AGBW02014900 OWR40914.1 DS231972 EDS29854.1 AJVK01016584 EDS29857.1 AJVK01016585 AJVK01016586 DS231896 EDS44436.1 GEZM01010812 JAV93814.1 EDS29860.1 GEZM01010817 GEZM01010816 JAV93809.1 GEZM01010815 JAV93810.1 JXUM01116018 KQ565940 KXJ70514.1 GEZM01010813 JAV93812.1 CH478228 EAT33496.1 GEZM01010811 JAV93815.1 AXCP01006762 GEZM01010814 JAV93811.1 APGK01038999 APGK01039000 KB740966 ENN76926.1 AXCN02001041 KB632186 ERL89696.1 KB632286 ERL91445.1 APGK01018199 APGK01018200 KB740071 ENN81698.1 CH963847 EDW73141.1 AAAB01008986 EAA00234.4 AXCM01007075 DS232244 EDS38311.1 APCN01003288 CAQQ02380437 AJWK01034404 AJWK01034405 ADMH02001044 ETN64334.1 ATLV01023896 KE525347 KFB49821.1 KYB25719.1 KQ971327 KYB28426.1 CH933809 EDW17776.1 CH477292 EAT44535.1 ATLV01023713 KE525345 KFB49419.1 JRES01000080 KNC34298.1 AE014296 AAF50555.3 CH954178 EDV50466.2 CH480817 EDW50846.1 CM002912 KMY98102.1 EDS38312.1 OUUW01000002 SPP77191.1 CH902618 EDV40708.1 AJWK01003969 AJWK01003970 AJWK01003971 AJWK01003972 KYB25718.1 ATLV01021410 KE525318 KFB46467.1 JXUM01054263 KQ561813 KXJ77445.1 CM000159 EDW93764.1 KQ971345 EFA05007.2 AXCM01001175
KOB71904.1 RSAL01000150 RVE45780.1 JTDY01002335 KOB71642.1 JTDY01005791 KOB66623.1 JTDY01008958 KOB64264.1 ODYU01012618 SOQ59034.1 NWSH01001201 PCG72148.1 KZ150309 PZC71475.1 KZ150082 PZC73838.1 KQ971362 EFA08873.2 KQ460597 KPJ13859.1 BABH01012830 ODYU01010562 SOQ55762.1 KQ459591 KPI97019.1 RSAL01000014 RVE53205.1 CVRI01000004 CRK87601.1 AGBW02014900 OWR40914.1 DS231972 EDS29854.1 AJVK01016584 EDS29857.1 AJVK01016585 AJVK01016586 DS231896 EDS44436.1 GEZM01010812 JAV93814.1 EDS29860.1 GEZM01010817 GEZM01010816 JAV93809.1 GEZM01010815 JAV93810.1 JXUM01116018 KQ565940 KXJ70514.1 GEZM01010813 JAV93812.1 CH478228 EAT33496.1 GEZM01010811 JAV93815.1 AXCP01006762 GEZM01010814 JAV93811.1 APGK01038999 APGK01039000 KB740966 ENN76926.1 AXCN02001041 KB632186 ERL89696.1 KB632286 ERL91445.1 APGK01018199 APGK01018200 KB740071 ENN81698.1 CH963847 EDW73141.1 AAAB01008986 EAA00234.4 AXCM01007075 DS232244 EDS38311.1 APCN01003288 CAQQ02380437 AJWK01034404 AJWK01034405 ADMH02001044 ETN64334.1 ATLV01023896 KE525347 KFB49821.1 KYB25719.1 KQ971327 KYB28426.1 CH933809 EDW17776.1 CH477292 EAT44535.1 ATLV01023713 KE525345 KFB49419.1 JRES01000080 KNC34298.1 AE014296 AAF50555.3 CH954178 EDV50466.2 CH480817 EDW50846.1 CM002912 KMY98102.1 EDS38312.1 OUUW01000002 SPP77191.1 CH902618 EDV40708.1 AJWK01003969 AJWK01003970 AJWK01003971 AJWK01003972 KYB25718.1 ATLV01021410 KE525318 KFB46467.1 JXUM01054263 KQ561813 KXJ77445.1 CM000159 EDW93764.1 KQ971345 EFA05007.2 AXCM01001175
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000007266
UP000053240
+ More
UP000053268 UP000183832 UP000007151 UP000002320 UP000092462 UP000069940 UP000249989 UP000008820 UP000075880 UP000192223 UP000075920 UP000019118 UP000075886 UP000030742 UP000075901 UP000007798 UP000007062 UP000075883 UP000075900 UP000075902 UP000076408 UP000075840 UP000076407 UP000075903 UP000075882 UP000075881 UP000015102 UP000092461 UP000069272 UP000075884 UP000095301 UP000095300 UP000000673 UP000030765 UP000192221 UP000009192 UP000037069 UP000000803 UP000008711 UP000001292 UP000268350 UP000007801 UP000002282 UP000075885
UP000053268 UP000183832 UP000007151 UP000002320 UP000092462 UP000069940 UP000249989 UP000008820 UP000075880 UP000192223 UP000075920 UP000019118 UP000075886 UP000030742 UP000075901 UP000007798 UP000007062 UP000075883 UP000075900 UP000075902 UP000076408 UP000075840 UP000076407 UP000075903 UP000075882 UP000075881 UP000015102 UP000092461 UP000069272 UP000075884 UP000095301 UP000095300 UP000000673 UP000030765 UP000192221 UP000009192 UP000037069 UP000000803 UP000008711 UP000001292 UP000268350 UP000007801 UP000002282 UP000075885
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEP9
A0A0L7LLW2
A0A0L7L972
A0A3S2LWT2
A0A0L7L821
A0A0L7KTM8
+ More
A0A0L7KMU4 A0A2H1X321 A0A2A4JK97 A0A2W1BBD0 A0A2W1BGB8 D6WXM6 A0A194R8M9 H9JHM1 A0A2H1WS18 A0A194PW83 A0A3S2NPU4 A0A1J1HMY9 A0A212EHF3 B0WKH8 A0A1B0DLX3 B0WKI1 A0A1B0DLX5 B0WDC3 A0A1Y1N7B1 B0WKI4 A0A1Y1N7X2 A0A1Y1NA93 A0A182H7C8 A0A1Y1N7C2 Q16GX7 A0A1Y1NB01 A0A182J6D0 A0A1Y1N7G4 A0A1S4G1A5 A0A1W4XR61 A0A182W972 N6U8E9 A0A182QS19 U4UGG3 U4UN62 N6TUG2 A0A182S9T8 B4ML83 Q7Q066 A0A182MLZ3 B0X0R9 A0A182R5F2 A0A182THH2 A0A182Y1U0 A0A182HQ19 A0A182XD77 A0A182UXK6 A0A182KSP4 A0A182K171 T1H5I3 A0A182IW77 A0A1B0GL64 A0A182WEP0 A0A182FU38 A0A182MYI9 A0A1I8M8B2 A0A1I8PG23 W5JNH9 A0A084WHX7 A0A139WCP8 A0A139WKH1 A0A1W4U6V2 B4KYR2 Q17DP0 A0A1I8Q829 A0A084WGS5 A0A0L0CPJ6 A0A1S4F6X7 Q9VS73 B3NFC4 B4HV75 A0A0J9RQW9 B0X0S0 A0A182JZW4 A0A182YLS3 A0A3B0JWU3 B3M5Z2 A0A1B0GH50 A0A139WCM4 A0A084W8C3 A0A182GCI9 B4PC79 A0A182FW18 A0A1I8NHK4 D2A5Z0 A0A182MGT5 A0A182PBQ5 A0A182KHA8 A0A182N248
A0A0L7KMU4 A0A2H1X321 A0A2A4JK97 A0A2W1BBD0 A0A2W1BGB8 D6WXM6 A0A194R8M9 H9JHM1 A0A2H1WS18 A0A194PW83 A0A3S2NPU4 A0A1J1HMY9 A0A212EHF3 B0WKH8 A0A1B0DLX3 B0WKI1 A0A1B0DLX5 B0WDC3 A0A1Y1N7B1 B0WKI4 A0A1Y1N7X2 A0A1Y1NA93 A0A182H7C8 A0A1Y1N7C2 Q16GX7 A0A1Y1NB01 A0A182J6D0 A0A1Y1N7G4 A0A1S4G1A5 A0A1W4XR61 A0A182W972 N6U8E9 A0A182QS19 U4UGG3 U4UN62 N6TUG2 A0A182S9T8 B4ML83 Q7Q066 A0A182MLZ3 B0X0R9 A0A182R5F2 A0A182THH2 A0A182Y1U0 A0A182HQ19 A0A182XD77 A0A182UXK6 A0A182KSP4 A0A182K171 T1H5I3 A0A182IW77 A0A1B0GL64 A0A182WEP0 A0A182FU38 A0A182MYI9 A0A1I8M8B2 A0A1I8PG23 W5JNH9 A0A084WHX7 A0A139WCP8 A0A139WKH1 A0A1W4U6V2 B4KYR2 Q17DP0 A0A1I8Q829 A0A084WGS5 A0A0L0CPJ6 A0A1S4F6X7 Q9VS73 B3NFC4 B4HV75 A0A0J9RQW9 B0X0S0 A0A182JZW4 A0A182YLS3 A0A3B0JWU3 B3M5Z2 A0A1B0GH50 A0A139WCM4 A0A084W8C3 A0A182GCI9 B4PC79 A0A182FW18 A0A1I8NHK4 D2A5Z0 A0A182MGT5 A0A182PBQ5 A0A182KHA8 A0A182N248
Ontologies
GO
PANTHER
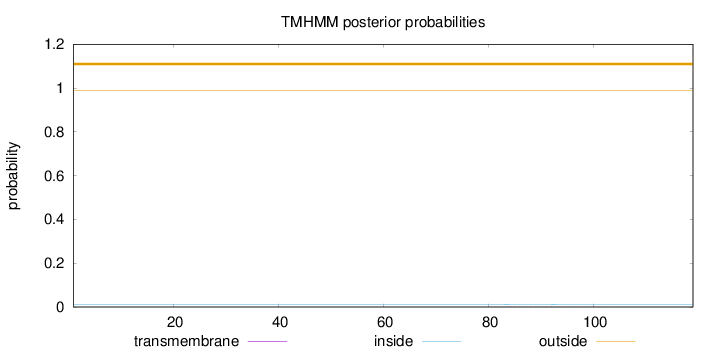
Topology
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00363
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01222
outside
1 - 119
Population Genetic Test Statistics
Pi
387.43164
Theta
202.157398
Tajima's D
2.956168
CLR
0.578337
CSRT
0.977401129943503
Interpretation
Uncertain
