Gene
KWMTBOMO04946
Annotation
PREDICTED:_putative_nuclease_HARBI1_[Acyrthosiphon_pisum]
Location in the cell
PlasmaMembrane Reliability : 2.25
Sequence
CDS
ATGAAGTCCTTGTATTTTGAATATTTGATCGGCACAACAACTCTGAGTGAAATAATTGAACACACATGCAAGAGTTTGTGGATATGTTTGCAACCTACATATATGCCTGAAAAAACAGAAGAGGATTGGATAAATATAGCAAATTTTTATTTTGAAAGAACACATTTTCCTAATTGTCTTGGTTCCATAGATGGAAAACATATACGAATAGAAAGGCCAGAAAATACTGGTTCAGAAGCCTTCAATTACAAAAAGTATTATTCTTTAATATTATTAGCTATAGCTGATGCAGACTATTGCTTTACTGCTATAGATGTTGGGGCATATGGATCGAATAGCGACTCTAGCGTTTTTAAAAACTCAAATTTTGGCAAAAGGTTTTTGAACAACCAAATGCATTTACCAGAATCTGCAACATTACCTGATTATGAGGAAGTCGGACCTTTACCGTTTGTTTTCGTAGGCGATGAAGCTTTTCCACTTATGGAGCACTTAATGAGGCCATATCCAAATAGAAATTTATCCATAAAACAAAGAGTTTATAACTATAGATTATCTTATGCCAGAAGGACTGTCGAATGTGCATTTGGGATCATGGCAAATAAATGGCGTATTTTACATAGACCATTAGATGTGAAATTAGATTTTTGCGACCATATTATTAAAGCTTGTTGCATATTGCACAACTTTGTAAGACAACATGACGGTAAACAGCTTAATGAATCGTTTCAAGTTTCAGAGATGCAGGGTATTGCTTCAATTCGTACGCGTTCTAGCAATAACGTTATAGATGTAAGAAATTTGTTTGCTGATTATTTTACATCTACTGCGGGCGCGTTGAGTTGGCAATATGAAAATATTTAA
Protein
MKSLYFEYLIGTTTLSEIIEHTCKSLWICLQPTYMPEKTEEDWINIANFYFERTHFPNCLGSIDGKHIRIERPENTGSEAFNYKKYYSLILLAIADADYCFTAIDVGAYGSNSDSSVFKNSNFGKRFLNNQMHLPESATLPDYEEVGPLPFVFVGDEAFPLMEHLMRPYPNRNLSIKQRVYNYRLSYARRTVECAFGIMANKWRILHRPLDVKLDFCDHIIKACCILHNFVRQHDGKQLNESFQVSEMQGIASIRTRSSNNVIDVRNLFADYFTSTAGALSWQYENI
Summary
Uniprot
J9K2X0
A0A2S2P1E6
A0A2S2NMG1
A0A3S2N8Q7
X1WRY0
A0A2S2NBK1
+ More
J9JIY0 J9M210 X1WMG6 A0A2J7QY78 A0A3S2P049 X1WIR3 J9K189 X1X279 A0A1B6KID6 J9K6X4 J9KI87 J9JMZ1 J9LWL5 A0A2S2P4H9 J9K876 J9JZA8 J9KD14 A0A1B6JBR0 A0A2S2P0U9 J9LWQ7 A0A2H8TIP0 A0A1E1WUI4 X1WIS8 X1WSE6 X1WZ84 A0A2H1VYY0 A0A2S2N8V8 X1WJJ7 J9KJZ3 A0A3S2TIS0 A0A2S2PAI8 J9K6E0 X1WXX7 A0A3S2L7Q6 J9LQ94 A0A2H8TVR0 A0A3S2L882 X1WKK4 S4NNW9 J9LKL3 A0A2S2PHY3 A0A0P4VZE0 X1XP58 A0A2J7PF61 X1WUC2 X1WQE4 A0A2G8JDA0 J9LS48 X1X0T9 J9K6X6 X1WPK0 A0A2S2PHB1 A0A3S2LPP0 A0A3S2N2Y6 X1XCN9 A0A0P4W2I2 B2GUF0 F6RCZ7 A0A0N0PET3 A0A2J7PD70 J9LAJ7 A0A0P4W5Q8 J9L7N5 A0A2J7QB64 A0A2J7PGM8 S4NZ68 A0A1W4X8I4 A0A147A2E9 A0A2J7PP30 A0A3P9KIC5 J9L3Z7 A0A2J7R0P9 A0A3P9K7T1 A0A1B6JTF8 A0A3B3QUT2 A0A3P9LX78 A0A3Q0QRU5 J9KFW1 A0A2J7R7I8 A0A2H1WK04 X1WIL6 A0A2G9RW26 A0A0N7ZBJ2 A0A2S2NUS4 A0A2S2NK21 J9L5G0 A1A649 A0A1B6HZT2 A0A1E1WI72 A0A3B3T7F3 A0A3S2LK83 A0A3Q1GW95 X1XF03 J9JQ73
J9JIY0 J9M210 X1WMG6 A0A2J7QY78 A0A3S2P049 X1WIR3 J9K189 X1X279 A0A1B6KID6 J9K6X4 J9KI87 J9JMZ1 J9LWL5 A0A2S2P4H9 J9K876 J9JZA8 J9KD14 A0A1B6JBR0 A0A2S2P0U9 J9LWQ7 A0A2H8TIP0 A0A1E1WUI4 X1WIS8 X1WSE6 X1WZ84 A0A2H1VYY0 A0A2S2N8V8 X1WJJ7 J9KJZ3 A0A3S2TIS0 A0A2S2PAI8 J9K6E0 X1WXX7 A0A3S2L7Q6 J9LQ94 A0A2H8TVR0 A0A3S2L882 X1WKK4 S4NNW9 J9LKL3 A0A2S2PHY3 A0A0P4VZE0 X1XP58 A0A2J7PF61 X1WUC2 X1WQE4 A0A2G8JDA0 J9LS48 X1X0T9 J9K6X6 X1WPK0 A0A2S2PHB1 A0A3S2LPP0 A0A3S2N2Y6 X1XCN9 A0A0P4W2I2 B2GUF0 F6RCZ7 A0A0N0PET3 A0A2J7PD70 J9LAJ7 A0A0P4W5Q8 J9L7N5 A0A2J7QB64 A0A2J7PGM8 S4NZ68 A0A1W4X8I4 A0A147A2E9 A0A2J7PP30 A0A3P9KIC5 J9L3Z7 A0A2J7R0P9 A0A3P9K7T1 A0A1B6JTF8 A0A3B3QUT2 A0A3P9LX78 A0A3Q0QRU5 J9KFW1 A0A2J7R7I8 A0A2H1WK04 X1WIL6 A0A2G9RW26 A0A0N7ZBJ2 A0A2S2NUS4 A0A2S2NK21 J9L5G0 A1A649 A0A1B6HZT2 A0A1E1WI72 A0A3B3T7F3 A0A3S2LK83 A0A3Q1GW95 X1XF03 J9JQ73
EMBL
ABLF02014609
GGMR01010668
MBY23287.1
GGMR01005523
MBY18142.1
RSAL01000782
+ More
RVE41156.1 ABLF02018117 GGMR01001869 MBY14488.1 ABLF02030395 ABLF02040883 ABLF02040911 ABLF02039417 NEVH01009123 PNF33550.1 RSAL01000073 RVE48993.1 ABLF02016720 ABLF02014546 ABLF02018112 ABLF02054180 ABLF02057104 GEBQ01028774 JAT11203.1 ABLF02010423 ABLF02036670 ABLF02023720 ABLF02023722 ABLF02049240 ABLF02035814 GGMR01011187 MBY23806.1 ABLF02013825 ABLF02039608 ABLF02029176 ABLF02029180 GECU01014635 GECU01011035 JAS93071.1 JAS96671.1 GGMR01010474 MBY23093.1 ABLF02010022 GFXV01002192 MBW13997.1 GDQN01000361 JAT90693.1 ABLF02010382 ABLF02017443 ABLF02042272 ABLF02053436 ABLF02063701 ABLF02058817 ODYU01005305 SOQ46028.1 GGMR01000966 MBY13585.1 ABLF02025001 ABLF02017112 RSAL01001047 RSAL01000133 RSAL01000110 RVE40968.1 RVE46314.1 RVE47157.1 GGMR01013773 MBY26392.1 ABLF02019581 ABLF02007334 RSAL01001281 RVE40853.1 ABLF02024888 GFXV01005837 MBW17642.1 RSAL01000093 RVE47949.1 ABLF02019551 GAIX01012104 JAA80456.1 ABLF02031827 GGMR01016408 MBY29027.1 GDRN01100681 JAI58521.1 ABLF02065969 NEVH01026085 PNF14965.1 ABLF02015607 ABLF02006842 ABLF02017285 ABLF02017286 ABLF02017287 ABLF02023975 ABLF02023979 ABLF02023981 ABLF02043231 ABLF02053316 ABLF02058285 MRZV01002436 PIK33713.1 ABLF02004403 ABLF02023972 ABLF02016114 ABLF02015081 GGMR01016212 MBY28831.1 RSAL01000893 RVE41064.1 RSAL01004553 RVE40126.1 ABLF02018875 ABLF02018876 ABLF02018877 GDRN01090017 JAI60545.1 BC166249 AAI66249.1 AAMC01113955 AAMC01113956 AAMC01113957 AAMC01113958 AAMC01113959 AAMC01113960 AAMC01113961 AAMC01113962 AAMC01113963 AAMC01113964 KQ459805 KPJ19882.1 NEVH01026393 PNF14277.1 ABLF02012792 GDRN01090013 JAI60547.1 ABLF02015257 ABLF02060247 NEVH01027118 NEVH01016302 NEVH01007440 NEVH01000280 PNF13625.1 PNF25824.1 PNF35689.1 PNF43673.1 NEVH01025174 PNF15485.1 GAIX01011467 JAA81093.1 GCES01013981 JAR72342.1 NEVH01023280 PNF18071.1 ABLF02013602 ABLF02013605 ABLF02053091 ABLF02066812 NEVH01008226 PNF34407.1 GECU01005239 JAT02468.1 NEVH01006731 PNF36798.1 ODYU01009172 SOQ53415.1 ABLF02017213 KV931512 PIO32080.1 GDRN01082635 JAI61800.1 GGMR01008300 MBY20919.1 GGMR01004653 MBY17272.1 ABLF02007889 ABLF02010298 BC128984 AAI28985.1 GECU01027536 JAS80170.1 GDQN01004369 JAT86685.1 RSAL01000083 RVE48487.1 ABLF02005780 ABLF02010250 ABLF02016429
RVE41156.1 ABLF02018117 GGMR01001869 MBY14488.1 ABLF02030395 ABLF02040883 ABLF02040911 ABLF02039417 NEVH01009123 PNF33550.1 RSAL01000073 RVE48993.1 ABLF02016720 ABLF02014546 ABLF02018112 ABLF02054180 ABLF02057104 GEBQ01028774 JAT11203.1 ABLF02010423 ABLF02036670 ABLF02023720 ABLF02023722 ABLF02049240 ABLF02035814 GGMR01011187 MBY23806.1 ABLF02013825 ABLF02039608 ABLF02029176 ABLF02029180 GECU01014635 GECU01011035 JAS93071.1 JAS96671.1 GGMR01010474 MBY23093.1 ABLF02010022 GFXV01002192 MBW13997.1 GDQN01000361 JAT90693.1 ABLF02010382 ABLF02017443 ABLF02042272 ABLF02053436 ABLF02063701 ABLF02058817 ODYU01005305 SOQ46028.1 GGMR01000966 MBY13585.1 ABLF02025001 ABLF02017112 RSAL01001047 RSAL01000133 RSAL01000110 RVE40968.1 RVE46314.1 RVE47157.1 GGMR01013773 MBY26392.1 ABLF02019581 ABLF02007334 RSAL01001281 RVE40853.1 ABLF02024888 GFXV01005837 MBW17642.1 RSAL01000093 RVE47949.1 ABLF02019551 GAIX01012104 JAA80456.1 ABLF02031827 GGMR01016408 MBY29027.1 GDRN01100681 JAI58521.1 ABLF02065969 NEVH01026085 PNF14965.1 ABLF02015607 ABLF02006842 ABLF02017285 ABLF02017286 ABLF02017287 ABLF02023975 ABLF02023979 ABLF02023981 ABLF02043231 ABLF02053316 ABLF02058285 MRZV01002436 PIK33713.1 ABLF02004403 ABLF02023972 ABLF02016114 ABLF02015081 GGMR01016212 MBY28831.1 RSAL01000893 RVE41064.1 RSAL01004553 RVE40126.1 ABLF02018875 ABLF02018876 ABLF02018877 GDRN01090017 JAI60545.1 BC166249 AAI66249.1 AAMC01113955 AAMC01113956 AAMC01113957 AAMC01113958 AAMC01113959 AAMC01113960 AAMC01113961 AAMC01113962 AAMC01113963 AAMC01113964 KQ459805 KPJ19882.1 NEVH01026393 PNF14277.1 ABLF02012792 GDRN01090013 JAI60547.1 ABLF02015257 ABLF02060247 NEVH01027118 NEVH01016302 NEVH01007440 NEVH01000280 PNF13625.1 PNF25824.1 PNF35689.1 PNF43673.1 NEVH01025174 PNF15485.1 GAIX01011467 JAA81093.1 GCES01013981 JAR72342.1 NEVH01023280 PNF18071.1 ABLF02013602 ABLF02013605 ABLF02053091 ABLF02066812 NEVH01008226 PNF34407.1 GECU01005239 JAT02468.1 NEVH01006731 PNF36798.1 ODYU01009172 SOQ53415.1 ABLF02017213 KV931512 PIO32080.1 GDRN01082635 JAI61800.1 GGMR01008300 MBY20919.1 GGMR01004653 MBY17272.1 ABLF02007889 ABLF02010298 BC128984 AAI28985.1 GECU01027536 JAS80170.1 GDQN01004369 JAT86685.1 RSAL01000083 RVE48487.1 ABLF02005780 ABLF02010250 ABLF02016429
Proteomes
ProteinModelPortal
J9K2X0
A0A2S2P1E6
A0A2S2NMG1
A0A3S2N8Q7
X1WRY0
A0A2S2NBK1
+ More
J9JIY0 J9M210 X1WMG6 A0A2J7QY78 A0A3S2P049 X1WIR3 J9K189 X1X279 A0A1B6KID6 J9K6X4 J9KI87 J9JMZ1 J9LWL5 A0A2S2P4H9 J9K876 J9JZA8 J9KD14 A0A1B6JBR0 A0A2S2P0U9 J9LWQ7 A0A2H8TIP0 A0A1E1WUI4 X1WIS8 X1WSE6 X1WZ84 A0A2H1VYY0 A0A2S2N8V8 X1WJJ7 J9KJZ3 A0A3S2TIS0 A0A2S2PAI8 J9K6E0 X1WXX7 A0A3S2L7Q6 J9LQ94 A0A2H8TVR0 A0A3S2L882 X1WKK4 S4NNW9 J9LKL3 A0A2S2PHY3 A0A0P4VZE0 X1XP58 A0A2J7PF61 X1WUC2 X1WQE4 A0A2G8JDA0 J9LS48 X1X0T9 J9K6X6 X1WPK0 A0A2S2PHB1 A0A3S2LPP0 A0A3S2N2Y6 X1XCN9 A0A0P4W2I2 B2GUF0 F6RCZ7 A0A0N0PET3 A0A2J7PD70 J9LAJ7 A0A0P4W5Q8 J9L7N5 A0A2J7QB64 A0A2J7PGM8 S4NZ68 A0A1W4X8I4 A0A147A2E9 A0A2J7PP30 A0A3P9KIC5 J9L3Z7 A0A2J7R0P9 A0A3P9K7T1 A0A1B6JTF8 A0A3B3QUT2 A0A3P9LX78 A0A3Q0QRU5 J9KFW1 A0A2J7R7I8 A0A2H1WK04 X1WIL6 A0A2G9RW26 A0A0N7ZBJ2 A0A2S2NUS4 A0A2S2NK21 J9L5G0 A1A649 A0A1B6HZT2 A0A1E1WI72 A0A3B3T7F3 A0A3S2LK83 A0A3Q1GW95 X1XF03 J9JQ73
J9JIY0 J9M210 X1WMG6 A0A2J7QY78 A0A3S2P049 X1WIR3 J9K189 X1X279 A0A1B6KID6 J9K6X4 J9KI87 J9JMZ1 J9LWL5 A0A2S2P4H9 J9K876 J9JZA8 J9KD14 A0A1B6JBR0 A0A2S2P0U9 J9LWQ7 A0A2H8TIP0 A0A1E1WUI4 X1WIS8 X1WSE6 X1WZ84 A0A2H1VYY0 A0A2S2N8V8 X1WJJ7 J9KJZ3 A0A3S2TIS0 A0A2S2PAI8 J9K6E0 X1WXX7 A0A3S2L7Q6 J9LQ94 A0A2H8TVR0 A0A3S2L882 X1WKK4 S4NNW9 J9LKL3 A0A2S2PHY3 A0A0P4VZE0 X1XP58 A0A2J7PF61 X1WUC2 X1WQE4 A0A2G8JDA0 J9LS48 X1X0T9 J9K6X6 X1WPK0 A0A2S2PHB1 A0A3S2LPP0 A0A3S2N2Y6 X1XCN9 A0A0P4W2I2 B2GUF0 F6RCZ7 A0A0N0PET3 A0A2J7PD70 J9LAJ7 A0A0P4W5Q8 J9L7N5 A0A2J7QB64 A0A2J7PGM8 S4NZ68 A0A1W4X8I4 A0A147A2E9 A0A2J7PP30 A0A3P9KIC5 J9L3Z7 A0A2J7R0P9 A0A3P9K7T1 A0A1B6JTF8 A0A3B3QUT2 A0A3P9LX78 A0A3Q0QRU5 J9KFW1 A0A2J7R7I8 A0A2H1WK04 X1WIL6 A0A2G9RW26 A0A0N7ZBJ2 A0A2S2NUS4 A0A2S2NK21 J9L5G0 A1A649 A0A1B6HZT2 A0A1E1WI72 A0A3B3T7F3 A0A3S2LK83 A0A3Q1GW95 X1XF03 J9JQ73
Ontologies
GO
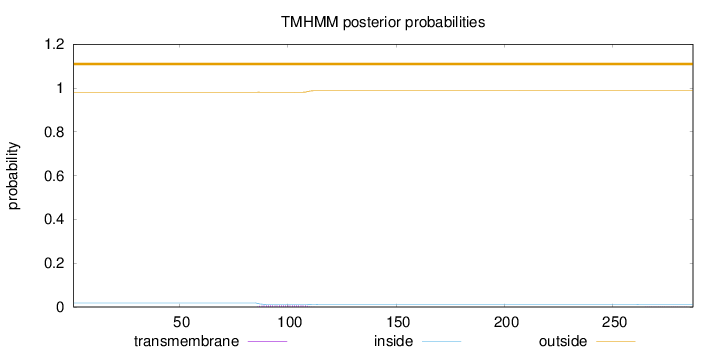
Topology
Length:
287
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18221
Exp number, first 60 AAs:
0.00115
Total prob of N-in:
0.01875
outside
1 - 287
Population Genetic Test Statistics
Pi
138.832696
Theta
147.657308
Tajima's D
0.010583
CLR
32.6107
CSRT
0.379031048447578
Interpretation
Uncertain
