Pre Gene Modal
BGIBMGA008001
Annotation
PREDICTED:_myc_box-dependent-interacting_protein_1_isoform_X1_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 2.689
Sequence
CDS
ATGGAAGCGATCACGGAGGTTTACGAGAGCGGCTGGTCAGGTCACGACCTGCTGTACGTCCAAGCTCAGAACATGGAGGTCCTTTGGCAGGACTTCTCGCACAAACTCGGCGACCAAGTACTCATACCGCTCAACACGTATACGAACCAATTCCCCGAAGTCAGGAAAAAAATCGAGAAACGTGGTCGCAAACTCGTAGACTACGACAGTCAAAGGCACAGCTTCCAGAACTTACAGGCGAATGCCGCTAAAAGAAGGGACGACGTAAAGGTGACGAAGGGACGCGAACAACTCGAAGAGGCCAAACGCACCTACGAAATATTGAACTCTGAACTGCACGACGAGCTGCCCGCGCTGTACGACTCCCGGGTATTGTTCTACGTCAACAACTTGCAAACACTCTTCTCCGCCGAGCAGGTCTTCCATTTCGAAAGCTCCAAGGTGTTCGCCGAACTAGAAGCGGTAACGGACAAGCTAGCGAACGACTGTCAGCGCGGCTCGTACACGAAGAAGGCGAACAACGTGTCCCCTCCCCCGCTGCCCGCGCTCCCCGCCGCCCCCGCGCACAACGGGAACGCCGCGCACACCAACACCGTGCAGACGCCCCCCTCCCCCGCGCTCAACGGCGACTCCCCCACGCTCTCCGGGAACAAAGAGTCGGCTCCGGTCAAAGAGACCGCACCGGACCCCAAAGACCCCGGAACGCCCGTCAAAGAGCCAGCTCCGGTTGCCAAAGAACCCGTATCGGTTGCCGAAGAACCGACGCCGATCAACAGCGAAACCGTTAAACCCGAGACTCCGCCCCCCACAATCGAAACGACCAATAGCAGCGAGCCCGCGACGGACAACAAAGTCGAACAGCTCTACGACATACCCGTCGGACCTACTACAGTCGTCGAGAAAATGAATAATAACGAAGAAACAAATAAACATACAAATGTCGAACAGAATTTAATCGAGAAGAACGAAAAGCGGAACAGTGAGAACGCCGAGGAGGTATACGACATACCAGTCGGAGCGACGCTAGCCGGCCTGCCGCCCGGCGTGCTGTACCGCGTGCGGGCCACATACCGGTACACGCGCGAGGATTCGGACGAGCTCTCGTTCGAAGTCGGCGACATCATACGCGTTGTCGAATACGACGATCCAGATGAACAGGAAGAGGGCTGGCTGATGGGCATCAAAGAAACGACCAATGAAAAAGGAATGTTCCCGGCGAACTTCACGCGGCCGATCTGA
Protein
MEAITEVYESGWSGHDLLYVQAQNMEVLWQDFSHKLGDQVLIPLNTYTNQFPEVRKKIEKRGRKLVDYDSQRHSFQNLQANAAKRRDDVKVTKGREQLEEAKRTYEILNSELHDELPALYDSRVLFYVNNLQTLFSAEQVFHFESSKVFAELEAVTDKLANDCQRGSYTKKANNVSPPPLPALPAAPAHNGNAAHTNTVQTPPSPALNGDSPTLSGNKESAPVKETAPDPKDPGTPVKEPAPVAKEPVSVAEEPTPINSETVKPETPPPTIETTNSSEPATDNKVEQLYDIPVGPTTVVEKMNNNEETNKHTNVEQNLIEKNEKRNSENAEEVYDIPVGATLAGLPPGVLYRVRATYRYTREDSDELSFEVGDIIRVVEYDDPDEQEEGWLMGIKETTNEKGMFPANFTRPI
Summary
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Uniprot
H9JEQ4
A0A2W1BKB7
A0A2H1WSY5
A0A3S2NNB4
A0A212EYU9
A0A194QEG6
+ More
A0A2A4JAE0 A0A194QK85 A0A2A3E7S0 A0A2J7PPD0 A0A2J7PP91 A0A195DTM8 A0A195EX96 A0A232F338 A0A026X0I2 E9IS54 A0A151I7D5 A0A2J7PPA8 A0A2J7PPA5 A0A195BKG1 V9ILS0 A0A0L7RDP0 A0A154PID6 E2AGE1 A0A0J7L7Z5 A0A151XGV3 A0A310S967 A0A2R7WXI0 E2BQJ7 A0A158P1D2 A0A1B6DR86 A0A1B6C6G8 A0A1Y1JX35 A0A2M4ACZ0 A0A146LH07 A0A0M8ZN15 V5H1R4 U4UBD3 T1IGD1 T1IVT4 A0A0P5X4J4 A0A0P4ZNJ3 A0A0P5BSZ4 A0A0P5ULS4 V5IIB5 A0A023G3A6 A0A0P4WBB6 A0A087ZWN6 A0A0P5BJS5 A0A0P5BPJ1 A0A0N8A4R9
A0A2A4JAE0 A0A194QK85 A0A2A3E7S0 A0A2J7PPD0 A0A2J7PP91 A0A195DTM8 A0A195EX96 A0A232F338 A0A026X0I2 E9IS54 A0A151I7D5 A0A2J7PPA8 A0A2J7PPA5 A0A195BKG1 V9ILS0 A0A0L7RDP0 A0A154PID6 E2AGE1 A0A0J7L7Z5 A0A151XGV3 A0A310S967 A0A2R7WXI0 E2BQJ7 A0A158P1D2 A0A1B6DR86 A0A1B6C6G8 A0A1Y1JX35 A0A2M4ACZ0 A0A146LH07 A0A0M8ZN15 V5H1R4 U4UBD3 T1IGD1 T1IVT4 A0A0P5X4J4 A0A0P4ZNJ3 A0A0P5BSZ4 A0A0P5ULS4 V5IIB5 A0A023G3A6 A0A0P4WBB6 A0A087ZWN6 A0A0P5BJS5 A0A0P5BPJ1 A0A0N8A4R9
Pubmed
EMBL
BABH01023471
KZ149993
PZC75522.1
ODYU01010779
SOQ56092.1
RSAL01000211
+ More
RVE44207.1 AGBW02011440 OWR46660.1 KQ459185 KPJ03340.1 NWSH01002337 PCG68494.1 KQ461198 KPJ05947.1 KZ288354 PBC27266.1 NEVH01023278 PNF18166.1 PNF18164.1 KQ980390 KYN16260.1 KQ981928 KYN32848.1 NNAY01001098 OXU25141.1 KK107046 EZA61782.1 GL765283 EFZ16673.1 KQ978415 KYM94072.1 PNF18168.1 PNF18165.1 KQ976449 KYM85813.1 JR050390 AEY61264.1 KQ414612 KOC69107.1 KQ434924 KZC11567.1 GL439310 EFN67474.1 LBMM01000307 KMR01438.1 KQ982138 KYQ59626.1 KQ763442 OAD55090.1 KK855863 PTY24171.1 GL449769 EFN82055.1 ADTU01006307 ADTU01006308 ADTU01006309 ADTU01006310 ADTU01006311 ADTU01006312 ADTU01006313 ADTU01006314 ADTU01006315 GEDC01009129 JAS28169.1 GEDC01028468 JAS08830.1 GEZM01098874 JAV53693.1 GGFK01005335 MBW38656.1 GDHC01019841 GDHC01016621 GDHC01011185 GDHC01003630 JAP98787.1 JAQ02008.1 JAQ07444.1 JAQ14999.1 KQ436019 KOX67565.1 GALX01001698 JAB66768.1 KB632184 ERL89643.1 ACPB03019049 JH431593 GDIP01077784 JAM25931.1 GDIP01213989 GDIP01180883 JAJ09413.1 GDIP01180884 JAJ42518.1 GDIP01128594 JAL75120.1 GANP01003477 JAB80991.1 GBBM01006737 JAC28681.1 GDRN01055674 JAI65946.1 GDIP01184038 JAJ39364.1 GDIP01182586 JAJ40816.1 GDIP01182585 JAJ40817.1
RVE44207.1 AGBW02011440 OWR46660.1 KQ459185 KPJ03340.1 NWSH01002337 PCG68494.1 KQ461198 KPJ05947.1 KZ288354 PBC27266.1 NEVH01023278 PNF18166.1 PNF18164.1 KQ980390 KYN16260.1 KQ981928 KYN32848.1 NNAY01001098 OXU25141.1 KK107046 EZA61782.1 GL765283 EFZ16673.1 KQ978415 KYM94072.1 PNF18168.1 PNF18165.1 KQ976449 KYM85813.1 JR050390 AEY61264.1 KQ414612 KOC69107.1 KQ434924 KZC11567.1 GL439310 EFN67474.1 LBMM01000307 KMR01438.1 KQ982138 KYQ59626.1 KQ763442 OAD55090.1 KK855863 PTY24171.1 GL449769 EFN82055.1 ADTU01006307 ADTU01006308 ADTU01006309 ADTU01006310 ADTU01006311 ADTU01006312 ADTU01006313 ADTU01006314 ADTU01006315 GEDC01009129 JAS28169.1 GEDC01028468 JAS08830.1 GEZM01098874 JAV53693.1 GGFK01005335 MBW38656.1 GDHC01019841 GDHC01016621 GDHC01011185 GDHC01003630 JAP98787.1 JAQ02008.1 JAQ07444.1 JAQ14999.1 KQ436019 KOX67565.1 GALX01001698 JAB66768.1 KB632184 ERL89643.1 ACPB03019049 JH431593 GDIP01077784 JAM25931.1 GDIP01213989 GDIP01180883 JAJ09413.1 GDIP01180884 JAJ42518.1 GDIP01128594 JAL75120.1 GANP01003477 JAB80991.1 GBBM01006737 JAC28681.1 GDRN01055674 JAI65946.1 GDIP01184038 JAJ39364.1 GDIP01182586 JAJ40816.1 GDIP01182585 JAJ40817.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000218220
UP000053240
+ More
UP000242457 UP000235965 UP000078492 UP000078541 UP000215335 UP000053097 UP000078542 UP000078540 UP000053825 UP000076502 UP000000311 UP000036403 UP000075809 UP000008237 UP000005205 UP000053105 UP000030742 UP000015103 UP000005203
UP000242457 UP000235965 UP000078492 UP000078541 UP000215335 UP000053097 UP000078542 UP000078540 UP000053825 UP000076502 UP000000311 UP000036403 UP000075809 UP000008237 UP000005205 UP000053105 UP000030742 UP000015103 UP000005203
PRIDE
Pfam
Interpro
IPR036028
SH3-like_dom_sf
+ More
IPR004148 BAR_dom
IPR027267 AH/BAR_dom_sf
IPR003005 Amphiphysin
IPR001452 SH3_domain
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR003603 U2A'_phosphoprotein32A_C
IPR003130 GED
IPR027417 P-loop_NTPase
IPR001849 PH_domain
IPR030381 G_DYNAMIN_dom
IPR011993 PH-like_dom_sf
IPR019762 Dynamin_GTPase_CS
IPR001401 Dynamin_GTPase
IPR027741 DNM1
IPR000375 Dynamin_central
IPR020850 GED_dom
IPR022812 Dynamin_SF
IPR004148 BAR_dom
IPR027267 AH/BAR_dom_sf
IPR003005 Amphiphysin
IPR001452 SH3_domain
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR003603 U2A'_phosphoprotein32A_C
IPR003130 GED
IPR027417 P-loop_NTPase
IPR001849 PH_domain
IPR030381 G_DYNAMIN_dom
IPR011993 PH-like_dom_sf
IPR019762 Dynamin_GTPase_CS
IPR001401 Dynamin_GTPase
IPR027741 DNM1
IPR000375 Dynamin_central
IPR020850 GED_dom
IPR022812 Dynamin_SF
Gene 3D
CDD
ProteinModelPortal
H9JEQ4
A0A2W1BKB7
A0A2H1WSY5
A0A3S2NNB4
A0A212EYU9
A0A194QEG6
+ More
A0A2A4JAE0 A0A194QK85 A0A2A3E7S0 A0A2J7PPD0 A0A2J7PP91 A0A195DTM8 A0A195EX96 A0A232F338 A0A026X0I2 E9IS54 A0A151I7D5 A0A2J7PPA8 A0A2J7PPA5 A0A195BKG1 V9ILS0 A0A0L7RDP0 A0A154PID6 E2AGE1 A0A0J7L7Z5 A0A151XGV3 A0A310S967 A0A2R7WXI0 E2BQJ7 A0A158P1D2 A0A1B6DR86 A0A1B6C6G8 A0A1Y1JX35 A0A2M4ACZ0 A0A146LH07 A0A0M8ZN15 V5H1R4 U4UBD3 T1IGD1 T1IVT4 A0A0P5X4J4 A0A0P4ZNJ3 A0A0P5BSZ4 A0A0P5ULS4 V5IIB5 A0A023G3A6 A0A0P4WBB6 A0A087ZWN6 A0A0P5BJS5 A0A0P5BPJ1 A0A0N8A4R9
A0A2A4JAE0 A0A194QK85 A0A2A3E7S0 A0A2J7PPD0 A0A2J7PP91 A0A195DTM8 A0A195EX96 A0A232F338 A0A026X0I2 E9IS54 A0A151I7D5 A0A2J7PPA8 A0A2J7PPA5 A0A195BKG1 V9ILS0 A0A0L7RDP0 A0A154PID6 E2AGE1 A0A0J7L7Z5 A0A151XGV3 A0A310S967 A0A2R7WXI0 E2BQJ7 A0A158P1D2 A0A1B6DR86 A0A1B6C6G8 A0A1Y1JX35 A0A2M4ACZ0 A0A146LH07 A0A0M8ZN15 V5H1R4 U4UBD3 T1IGD1 T1IVT4 A0A0P5X4J4 A0A0P4ZNJ3 A0A0P5BSZ4 A0A0P5ULS4 V5IIB5 A0A023G3A6 A0A0P4WBB6 A0A087ZWN6 A0A0P5BJS5 A0A0P5BPJ1 A0A0N8A4R9
PDB
1URU
E-value=2.90022e-66,
Score=640
Ontologies
GO
PANTHER
Topology
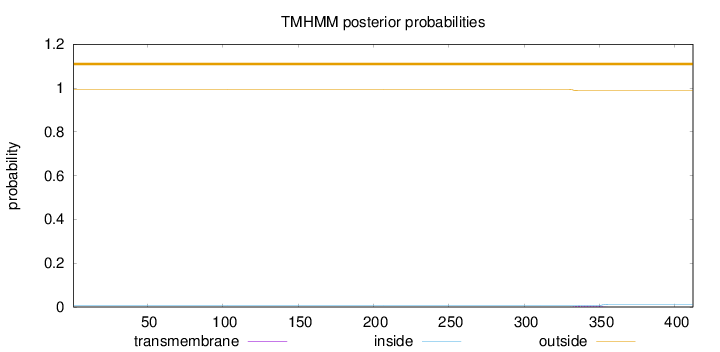
Length:
412
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10418
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00734
outside
1 - 412
Population Genetic Test Statistics
Pi
204.799771
Theta
194.282516
Tajima's D
0.103502
CLR
158.383989
CSRT
0.394630268486576
Interpretation
Uncertain
