Gene
KWMTBOMO04936
Pre Gene Modal
BGIBMGA008002
Annotation
PREDICTED:_fatty-acid_amide_hydrolase_2-A-like_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.578 Mitochondrial Reliability : 1.506
Sequence
CDS
ATGGAGGTTGGTGTACGGCTGGTGGGTATACTGCTGCGGTTCCTGAACTTCATTCTCACTCCTTTGTTCTGGTTAAAGTCGCACAAGAGGACGTGCGTTCCTCCCGTAACGGAGCCCATGCTGTTCAAAGGCGCGGCCGACTTGGCACGCGAAATTCGCAATGGCGAGATCACGAGTACACAAGTTGTGACTATGTACATTAAACGTATAAAGGAAGTGAATCCGTACTTGAATGCGGTTGTGGAGGATCGCTACAGAGCTGCCCTCTACGAAGCGAGGGCTTGTGACGTACAGATCAAAGAAGCGATGCGACTTGGCACCATAGAAGACTTAGTCAAAAACAAACCTCTACTGGGAGTCCCTTTTACTGTGAAAGAGAGCTGCTCTCTGGGAGGCCTCTCAAATTCCGTCGGTTGTCTGGAGAACGCAGGACGGAAAGCGACTGCTGACGGCGCGGCTGTGGCTCTTGTGAGGGACGCGGGGGCCATCCCGCTCCTCGTGTCCAACACTCCGGAGCTGTGTCTCGGCTGGGAGACTACGAACCTACTAACAGGCACTACTAATAACCCGTATTGGCTCAGCAAGACTCCGGGCGGATCTTCTGGTGGCGAGGCCGCATTACTAGCTAGCGCGGCGTCTCCTTTCAGTGTCTCTTCCGACATCGCGGGATCCATTCGCGTTCCCGCCGCGTTCTGCGGCGTCTATGGACACAAACCTACTCCAGGTATCATCCCTATAGAGGGTCACATTCCGACTCTAAGCGATGAGCAGTTTCCAAAGTTCCTGACGGTGGGCCCCATGGCGAGGAAGGCTGAAGACCTTCCCTTGTTGTTGAACGTGATGGCGGGCGAGAACCGCCACGTTCTGAACCTCGACGAACCCGTCAACATCGAAGACATTGAGGTGTTCTACATGACGGAAGCCATGGACTCCATAGCCCTGATCAAAGTTGACAAGTGTATCCAAGACGCCGTGCTGGAGGCCGTTCAATACCTGAGTCGATGTGGAGCCACCGTCAGCGAGGAAAAATTCAAAGGCTTAGAAGATTCAGTCGAAATATCGATATCCGTGTTTTTCTCGATGAAGGACATCCCGAATATGCTGCAGGATCCGGCCGATCCGAAGCGTGAAAAAAATTTGTTTTTAGAAATACTTAAGTGTCCCATCGGCGGAGCGTCGCGTTCGTTACAAGGCCTCAGCTTCGCCTTGATAGAGAAGACGCGGCTCTTCATACCGAAGTCGAAAACGGACTTCTACAAAGACAAGGCGGACAAACTCCGGACCGAGATCACGGTGCGTGAAGCTTTTACCACGATGTATGAGAATTGA
Protein
MEVGVRLVGILLRFLNFILTPLFWLKSHKRTCVPPVTEPMLFKGAADLAREIRNGEITSTQVVTMYIKRIKEVNPYLNAVVEDRYRAALYEARACDVQIKEAMRLGTIEDLVKNKPLLGVPFTVKESCSLGGLSNSVGCLENAGRKATADGAAVALVRDAGAIPLLVSNTPELCLGWETTNLLTGTTNNPYWLSKTPGGSSGGEAALLASAASPFSVSSDIAGSIRVPAAFCGVYGHKPTPGIIPIEGHIPTLSDEQFPKFLTVGPMARKAEDLPLLLNVMAGENRHVLNLDEPVNIEDIEVFYMTEAMDSIALIKVDKCIQDAVLEAVQYLSRCGATVSEEKFKGLEDSVEISISVFFSMKDIPNMLQDPADPKREKNLFLEILKCPIGGASRSLQGLSFALIEKTRLFIPKSKTDFYKDKADKLRTEITVREAFTTMYEN
Summary
Uniprot
H9JEQ5
A0A2A4J2Y4
A0A2H1VC64
A0A2W1BMK1
A0A212EYT2
A0A2J7PRL0
+ More
A0A067R7C9 A0A2J7PRK9 A0A1B6I2Q0 A0A1B6EZP8 A0A1B6DIM6 A0A224XN95 A0A0V0GCT0 A0A1B6L425 A0A182IVS1 A0A084W1M1 U5ELF6 A0A182QEG0 A0A336LYA9 Q5TTQ3 A0A182UP47 A0A182LFD6 A0A023ETZ1 A0A0P4VSF5 Q16GJ2 T1HPU6 A0A182GJX7 Q16UE9 A0A336LUQ5 R4WS00 A0A182W2W7 A0A182KDG7 A0A1B6I5N9 A0A182TWL3 A0A2M4ATP5 A0A2M4ATM8 A0A2M4AT49 A0A2M4AT89 A0A182PA47 T1DT07 A0A182HT95 A0A2M4BL10 A0A2M4BLU7 A0A0N7Z9H6 T1HQS6 A0A182RYG1 D6W8X8 A0A1J1IQM9 A0A182F8B7 A0A182XIA9 A0A182NA72 A0A1B6L1N5 B4M152 A0A0L7QQF4 A0A158NHU3 A0A182Y777 Q29AS7 B4G604 A0A1B6C6F5 W5JQ74 A0A182MVM3 A0A1B6E1G0 A0A195FEN1 V5GZB7 A0A3B0JPY9 A0A1W4VR76 B4PSK8 B3LZ88 B0W6F2 A0A195EGA3 B4K971 A0A1Q3FLI7 A0A1Q3FKP7 A0A1Q3FLF2 A0A067R9Z7 B4JF59 B4HM32 B3NZX5 A0A0C9RH59 B4QYQ7 A0A2M4DQN3 A0A151WRH2 Q9VHW0 A0A2P8YWS8 F4WYB1 B3LV58 A0A0A9ZDX3 A0A0A9X9N1 F4WG47 A0A1W4W2Q1 S4NRR4 A0A1B6EPA5 A0A084W1M0 A0A1I8NQB7 A0A195CD02 A0A0M4F363 A0A182J2R6 B4JF58 T1PB28
A0A067R7C9 A0A2J7PRK9 A0A1B6I2Q0 A0A1B6EZP8 A0A1B6DIM6 A0A224XN95 A0A0V0GCT0 A0A1B6L425 A0A182IVS1 A0A084W1M1 U5ELF6 A0A182QEG0 A0A336LYA9 Q5TTQ3 A0A182UP47 A0A182LFD6 A0A023ETZ1 A0A0P4VSF5 Q16GJ2 T1HPU6 A0A182GJX7 Q16UE9 A0A336LUQ5 R4WS00 A0A182W2W7 A0A182KDG7 A0A1B6I5N9 A0A182TWL3 A0A2M4ATP5 A0A2M4ATM8 A0A2M4AT49 A0A2M4AT89 A0A182PA47 T1DT07 A0A182HT95 A0A2M4BL10 A0A2M4BLU7 A0A0N7Z9H6 T1HQS6 A0A182RYG1 D6W8X8 A0A1J1IQM9 A0A182F8B7 A0A182XIA9 A0A182NA72 A0A1B6L1N5 B4M152 A0A0L7QQF4 A0A158NHU3 A0A182Y777 Q29AS7 B4G604 A0A1B6C6F5 W5JQ74 A0A182MVM3 A0A1B6E1G0 A0A195FEN1 V5GZB7 A0A3B0JPY9 A0A1W4VR76 B4PSK8 B3LZ88 B0W6F2 A0A195EGA3 B4K971 A0A1Q3FLI7 A0A1Q3FKP7 A0A1Q3FLF2 A0A067R9Z7 B4JF59 B4HM32 B3NZX5 A0A0C9RH59 B4QYQ7 A0A2M4DQN3 A0A151WRH2 Q9VHW0 A0A2P8YWS8 F4WYB1 B3LV58 A0A0A9ZDX3 A0A0A9X9N1 F4WG47 A0A1W4W2Q1 S4NRR4 A0A1B6EPA5 A0A084W1M0 A0A1I8NQB7 A0A195CD02 A0A0M4F363 A0A182J2R6 B4JF58 T1PB28
Pubmed
19121390
28756777
22118469
24845553
24438588
12364791
+ More
14747013 17210077 20966253 24945155 27129103 17510324 26483478 23691247 18362917 19820115 17994087 21347285 25244985 15632085 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 21719571 25401762 26823975 23622113
14747013 17210077 20966253 24945155 27129103 17510324 26483478 23691247 18362917 19820115 17994087 21347285 25244985 15632085 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 21719571 25401762 26823975 23622113
EMBL
BABH01023471
NWSH01003855
PCG65753.1
ODYU01001772
SOQ38438.1
KZ149993
+ More
PZC75521.1 AGBW02011440 OWR46659.1 NEVH01021956 PNF18972.1 KK852827 KDR15405.1 PNF18975.1 GECU01026504 JAS81202.1 GECZ01026297 JAS43472.1 GEDC01011786 GEDC01007189 JAS25512.1 JAS30109.1 GFTR01006935 JAW09491.1 GECL01000951 JAP05173.1 GEBQ01021485 JAT18492.1 ATLV01019387 KE525269 KFB44115.1 GANO01001426 JAB58445.1 AXCN02000218 UFQT01000202 SSX21663.1 AAAB01008859 EAL40937.3 GAPW01001232 JAC12366.1 GDKW01001082 JAI55513.1 CH478271 EAT33361.1 ACPB03008263 JXUM01013060 JXUM01013061 KQ560367 KXJ82787.1 CH477623 EAT38148.1 SSX21664.1 AK417477 BAN20692.1 GECU01025491 JAS82215.1 GGFK01010848 MBW44169.1 GGFK01010825 MBW44146.1 GGFK01010623 MBW43944.1 GGFK01010672 MBW43993.1 GAMD01001244 JAB00347.1 APCN01000423 APCN01000424 GGFJ01004616 MBW53757.1 GGFJ01004617 MBW53758.1 GDKW01000523 JAI56072.1 ACPB03027148 KQ971312 EEZ98237.1 CVRI01000057 CRL02527.1 GEBQ01022379 JAT17598.1 CH940650 EDW67463.1 KQ414786 KOC60848.1 ADTU01016061 ADTU01016062 ADTU01016063 CM000070 EAL27273.2 CH479179 EDW23756.1 GEDC01028212 JAS09086.1 ADMH02000651 ETN65438.1 AXCM01000066 GEDC01005536 JAS31762.1 KQ981636 KYN38836.1 GALX01002628 JAB65838.1 OUUW01000007 SPP82973.1 CM000160 EDW96468.1 CH902617 EDV43015.1 DS231848 EDS36626.1 KQ978957 KYN27268.1 CH933806 EDW14484.1 GFDL01006653 JAV28392.1 GFDL01006886 JAV28159.1 GFDL01006729 JAV28316.1 KDR15404.1 CH916369 EDV93340.1 CH480815 EDW43080.1 CH954181 EDV49973.1 GBYB01006266 JAG76033.1 CM000364 EDX13815.1 GGFL01015651 MBW79829.1 KQ982813 KYQ50275.1 AE014297 BT021469 AAF54189.1 AAX33617.1 PYGN01000313 PSN48701.1 GL888439 EGI60868.1 EDV42530.1 GBHO01029753 GBHO01012549 GBHO01002069 GBHO01002065 GBHO01002061 GBRD01006321 GBRD01006320 GBRD01006319 GBRD01006318 GBRD01006317 GBRD01006316 GDHC01019820 GDHC01006937 JAG13851.1 JAG31055.1 JAG41535.1 JAG41539.1 JAG41543.1 JAG59500.1 JAP98808.1 JAQ11692.1 GBHO01029794 GBHO01012550 GBHO01002070 JAG13810.1 JAG31054.1 JAG41534.1 GL888128 EGI66814.1 GAIX01014342 JAA78218.1 GECZ01030096 JAS39673.1 KFB44114.1 KQ978023 KYM98096.1 CP012526 ALC45620.1 EDV93339.1 KA645163 AFP59792.1
PZC75521.1 AGBW02011440 OWR46659.1 NEVH01021956 PNF18972.1 KK852827 KDR15405.1 PNF18975.1 GECU01026504 JAS81202.1 GECZ01026297 JAS43472.1 GEDC01011786 GEDC01007189 JAS25512.1 JAS30109.1 GFTR01006935 JAW09491.1 GECL01000951 JAP05173.1 GEBQ01021485 JAT18492.1 ATLV01019387 KE525269 KFB44115.1 GANO01001426 JAB58445.1 AXCN02000218 UFQT01000202 SSX21663.1 AAAB01008859 EAL40937.3 GAPW01001232 JAC12366.1 GDKW01001082 JAI55513.1 CH478271 EAT33361.1 ACPB03008263 JXUM01013060 JXUM01013061 KQ560367 KXJ82787.1 CH477623 EAT38148.1 SSX21664.1 AK417477 BAN20692.1 GECU01025491 JAS82215.1 GGFK01010848 MBW44169.1 GGFK01010825 MBW44146.1 GGFK01010623 MBW43944.1 GGFK01010672 MBW43993.1 GAMD01001244 JAB00347.1 APCN01000423 APCN01000424 GGFJ01004616 MBW53757.1 GGFJ01004617 MBW53758.1 GDKW01000523 JAI56072.1 ACPB03027148 KQ971312 EEZ98237.1 CVRI01000057 CRL02527.1 GEBQ01022379 JAT17598.1 CH940650 EDW67463.1 KQ414786 KOC60848.1 ADTU01016061 ADTU01016062 ADTU01016063 CM000070 EAL27273.2 CH479179 EDW23756.1 GEDC01028212 JAS09086.1 ADMH02000651 ETN65438.1 AXCM01000066 GEDC01005536 JAS31762.1 KQ981636 KYN38836.1 GALX01002628 JAB65838.1 OUUW01000007 SPP82973.1 CM000160 EDW96468.1 CH902617 EDV43015.1 DS231848 EDS36626.1 KQ978957 KYN27268.1 CH933806 EDW14484.1 GFDL01006653 JAV28392.1 GFDL01006886 JAV28159.1 GFDL01006729 JAV28316.1 KDR15404.1 CH916369 EDV93340.1 CH480815 EDW43080.1 CH954181 EDV49973.1 GBYB01006266 JAG76033.1 CM000364 EDX13815.1 GGFL01015651 MBW79829.1 KQ982813 KYQ50275.1 AE014297 BT021469 AAF54189.1 AAX33617.1 PYGN01000313 PSN48701.1 GL888439 EGI60868.1 EDV42530.1 GBHO01029753 GBHO01012549 GBHO01002069 GBHO01002065 GBHO01002061 GBRD01006321 GBRD01006320 GBRD01006319 GBRD01006318 GBRD01006317 GBRD01006316 GDHC01019820 GDHC01006937 JAG13851.1 JAG31055.1 JAG41535.1 JAG41539.1 JAG41543.1 JAG59500.1 JAP98808.1 JAQ11692.1 GBHO01029794 GBHO01012550 GBHO01002070 JAG13810.1 JAG31054.1 JAG41534.1 GL888128 EGI66814.1 GAIX01014342 JAA78218.1 GECZ01030096 JAS39673.1 KFB44114.1 KQ978023 KYM98096.1 CP012526 ALC45620.1 EDV93339.1 KA645163 AFP59792.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000235965
UP000027135
UP000075880
+ More
UP000030765 UP000075886 UP000007062 UP000075903 UP000075882 UP000008820 UP000015103 UP000069940 UP000249989 UP000075920 UP000075881 UP000075902 UP000075885 UP000075840 UP000075900 UP000007266 UP000183832 UP000069272 UP000076407 UP000075884 UP000008792 UP000053825 UP000005205 UP000076408 UP000001819 UP000008744 UP000000673 UP000075883 UP000078541 UP000268350 UP000192221 UP000002282 UP000007801 UP000002320 UP000078492 UP000009192 UP000001070 UP000001292 UP000008711 UP000000304 UP000075809 UP000000803 UP000245037 UP000007755 UP000095300 UP000078542 UP000092553
UP000030765 UP000075886 UP000007062 UP000075903 UP000075882 UP000008820 UP000015103 UP000069940 UP000249989 UP000075920 UP000075881 UP000075902 UP000075885 UP000075840 UP000075900 UP000007266 UP000183832 UP000069272 UP000076407 UP000075884 UP000008792 UP000053825 UP000005205 UP000076408 UP000001819 UP000008744 UP000000673 UP000075883 UP000078541 UP000268350 UP000192221 UP000002282 UP000007801 UP000002320 UP000078492 UP000009192 UP000001070 UP000001292 UP000008711 UP000000304 UP000075809 UP000000803 UP000245037 UP000007755 UP000095300 UP000078542 UP000092553
Interpro
SUPFAM
SSF75304
SSF75304
Gene 3D
ProteinModelPortal
H9JEQ5
A0A2A4J2Y4
A0A2H1VC64
A0A2W1BMK1
A0A212EYT2
A0A2J7PRL0
+ More
A0A067R7C9 A0A2J7PRK9 A0A1B6I2Q0 A0A1B6EZP8 A0A1B6DIM6 A0A224XN95 A0A0V0GCT0 A0A1B6L425 A0A182IVS1 A0A084W1M1 U5ELF6 A0A182QEG0 A0A336LYA9 Q5TTQ3 A0A182UP47 A0A182LFD6 A0A023ETZ1 A0A0P4VSF5 Q16GJ2 T1HPU6 A0A182GJX7 Q16UE9 A0A336LUQ5 R4WS00 A0A182W2W7 A0A182KDG7 A0A1B6I5N9 A0A182TWL3 A0A2M4ATP5 A0A2M4ATM8 A0A2M4AT49 A0A2M4AT89 A0A182PA47 T1DT07 A0A182HT95 A0A2M4BL10 A0A2M4BLU7 A0A0N7Z9H6 T1HQS6 A0A182RYG1 D6W8X8 A0A1J1IQM9 A0A182F8B7 A0A182XIA9 A0A182NA72 A0A1B6L1N5 B4M152 A0A0L7QQF4 A0A158NHU3 A0A182Y777 Q29AS7 B4G604 A0A1B6C6F5 W5JQ74 A0A182MVM3 A0A1B6E1G0 A0A195FEN1 V5GZB7 A0A3B0JPY9 A0A1W4VR76 B4PSK8 B3LZ88 B0W6F2 A0A195EGA3 B4K971 A0A1Q3FLI7 A0A1Q3FKP7 A0A1Q3FLF2 A0A067R9Z7 B4JF59 B4HM32 B3NZX5 A0A0C9RH59 B4QYQ7 A0A2M4DQN3 A0A151WRH2 Q9VHW0 A0A2P8YWS8 F4WYB1 B3LV58 A0A0A9ZDX3 A0A0A9X9N1 F4WG47 A0A1W4W2Q1 S4NRR4 A0A1B6EPA5 A0A084W1M0 A0A1I8NQB7 A0A195CD02 A0A0M4F363 A0A182J2R6 B4JF58 T1PB28
A0A067R7C9 A0A2J7PRK9 A0A1B6I2Q0 A0A1B6EZP8 A0A1B6DIM6 A0A224XN95 A0A0V0GCT0 A0A1B6L425 A0A182IVS1 A0A084W1M1 U5ELF6 A0A182QEG0 A0A336LYA9 Q5TTQ3 A0A182UP47 A0A182LFD6 A0A023ETZ1 A0A0P4VSF5 Q16GJ2 T1HPU6 A0A182GJX7 Q16UE9 A0A336LUQ5 R4WS00 A0A182W2W7 A0A182KDG7 A0A1B6I5N9 A0A182TWL3 A0A2M4ATP5 A0A2M4ATM8 A0A2M4AT49 A0A2M4AT89 A0A182PA47 T1DT07 A0A182HT95 A0A2M4BL10 A0A2M4BLU7 A0A0N7Z9H6 T1HQS6 A0A182RYG1 D6W8X8 A0A1J1IQM9 A0A182F8B7 A0A182XIA9 A0A182NA72 A0A1B6L1N5 B4M152 A0A0L7QQF4 A0A158NHU3 A0A182Y777 Q29AS7 B4G604 A0A1B6C6F5 W5JQ74 A0A182MVM3 A0A1B6E1G0 A0A195FEN1 V5GZB7 A0A3B0JPY9 A0A1W4VR76 B4PSK8 B3LZ88 B0W6F2 A0A195EGA3 B4K971 A0A1Q3FLI7 A0A1Q3FKP7 A0A1Q3FLF2 A0A067R9Z7 B4JF59 B4HM32 B3NZX5 A0A0C9RH59 B4QYQ7 A0A2M4DQN3 A0A151WRH2 Q9VHW0 A0A2P8YWS8 F4WYB1 B3LV58 A0A0A9ZDX3 A0A0A9X9N1 F4WG47 A0A1W4W2Q1 S4NRR4 A0A1B6EPA5 A0A084W1M0 A0A1I8NQB7 A0A195CD02 A0A0M4F363 A0A182J2R6 B4JF58 T1PB28
PDB
5H6T
E-value=8.29046e-23,
Score=265
Ontologies
Topology
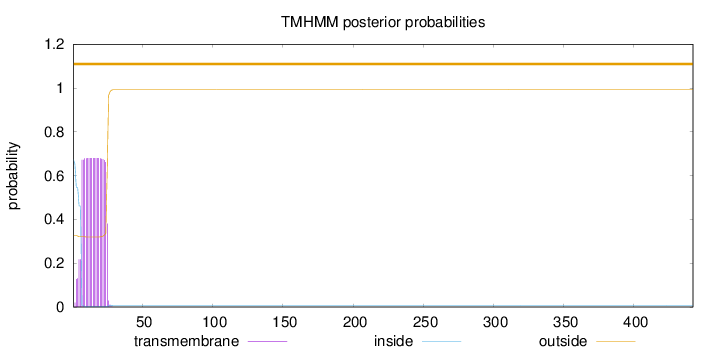
Length:
442
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.3226999999999
Exp number, first 60 AAs:
13.31996
Total prob of N-in:
0.67321
POSSIBLE N-term signal
sequence
outside
1 - 442
Population Genetic Test Statistics
Pi
188.959959
Theta
184.377506
Tajima's D
0.983393
CLR
0.205127
CSRT
0.656817159142043
Interpretation
Uncertain
