Gene
KWMTBOMO04933 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008076
Annotation
PREDICTED:_2-oxoisovalerate_dehydrogenase_subunit_beta?_mitochondrial_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.256 Mitochondrial Reliability : 1.58
Sequence
CDS
ATGACAAGTTTAAGTACAAAAAGTATTGTGTTCCATAAATTAATTCGAAATGTTAATAACTACGCTAAAAGAATGTCTTCACATTTTATTTATTATCCAGATAAAGAAAGGCCAGTAGATGGAGAAACGACAAAGATGAATATGATGCAAGCAATAAACAATGCAATGGACATCACATTGAAGAATAATCCAACTGCTGTATTGTTCGGTGAAGATGTCGCATTCGGGGGTGTCTTCAGATGTGCTTTGGGGTTACAGGAGAAATATGGCAAGGACCGTGTTTTCAATACTCCACTATGTGAACAGGGTATTGCCGGCTTCGGTATAGGTCTAGCCACTGCAGGCGCAACTGCTATTGCTGAAATACAATTTGCTGATTACATATTTCCAGCATTTGATCAGATAGTGAACGAAGCTGCCAAAGCGAGGTACAGATCTGGAGGGGAGTACGACAGCGGCGCCCTCACGGTCCGCGCGCCCTGCAGTGCGGTCGGGCACGGCGGACTCTACCACTCCCAGAGTCCGGAGGCCTTCTTTGCACATGTCCCCGGTCTCCGGGTGGTGGTGCCTCGCGGGCCCATAGCGGCCAAGGGTCTGCTGCTGGCCTGCATCCGCGAGAGGGACCCCTGCGTGTTCCTGGAACCGAAGATACTGTACCGGTCGGCAGCCGAAGAAGTACCCGTCGAGGATTACACGCTACCGCTGGGAAAGGCGCAGACGTTAAGAGTCGGCGCCGCGGCCACGCTGGTGGGCTGGGGCACGCAGGTTCACGTGCTGCTGGAGGTAGCAGACATGGCGCGGGACAAGCTCGGCGTCAGCTGTGACGTCATCGATCTGCAGTCGATCCTGCCCTGGGACGAGGAGACCGTGTGCAATTCCGTGAAGAAAACTGGGCGGTGCTTGATATCCCACGAGGCCCCGCTCACGTCGGGGTTCGGCGCCGAGCTCGCCGCCACCGTGCAGGAGGAATGTTTTCTGCACTTGGAGGCGCCGATAGCTCGAGTGACCGGCTGGGACGCGCCCTTCCCGCATGTCTTCGAACCTTTCTACTTACCGGACAAGTGGCGCTGCTACCAAGCCTTGATACAACTTATTAACTATTGA
Protein
MTSLSTKSIVFHKLIRNVNNYAKRMSSHFIYYPDKERPVDGETTKMNMMQAINNAMDITLKNNPTAVLFGEDVAFGGVFRCALGLQEKYGKDRVFNTPLCEQGIAGFGIGLATAGATAIAEIQFADYIFPAFDQIVNEAAKARYRSGGEYDSGALTVRAPCSAVGHGGLYHSQSPEAFFAHVPGLRVVVPRGPIAAKGLLLACIRERDPCVFLEPKILYRSAAEEVPVEDYTLPLGKAQTLRVGAAATLVGWGTQVHVLLEVADMARDKLGVSCDVIDLQSILPWDEETVCNSVKKTGRCLISHEAPLTSGFGAELAATVQEECFLHLEAPIARVTGWDAPFPHVFEPFYLPDKWRCYQALIQLINY
Summary
Uniprot
A0A2A4JEM1
A0A2H1VC69
A0A3S2LUE1
A0A194QLW2
H9JEX9
A0A212ELF7
+ More
A0A194QD47 B0X5D4 A0A1Q3F6Y1 A0A0N8ES10 A0A182GQD9 A0A182TMH6 A0A182WJ45 A0A2M3Z7D0 A0A2M4AL73 A0A2M4AL59 A0A2M4AM84 A0A182PQB0 Q7QJI1 A0A2M3Z7P6 A0A182X3L8 A0A182I3F1 A0A2M4BRR3 A0A084WNF8 A0A2M3Z7U5 A0A182NHR4 A0A182JNC6 W5JJS3 A0A182RNF4 D6W8S4 A0A182M803 A0A182KRH0 A0A2R5L8N2 A0A182VFT1 A0A182QIR8 A0A182FG02 G3MQQ0 A0A182JD95 A0A023GME0 A0A182XZ02 A0A1Y1LXF5 U5ESH2 E9HG12 A0A226CZB8 A0A1E1X607 A0A0P4ZZY0 A0A0L7QJJ9 A0A0P6I319 A0A088ADW9 A0A0P6F5F7 A0A131XLK9 A0A0P6AHU0 L7MB32 A0A224Z7A2 A0A0A9XMT4 A0A023F899 A0A023FX40 A0A0P6EUA5 A0A1J1J880 A0A1B6DYD4 A0A224XEG4 R4G3S1 A0A131Z0J0 A0A1L8E144 A0A0N7Z9G1 T1KER6 A0A3P9I8H2 A0A3P9JWQ4 A0A3B3D2S8 A0A232EVA3 A0A0P5MZL2 A0A0L0CJM0 A0A0P5KRP3 A0A3B3HQ15 A0A0P5VTF6 A0A0T6BBV5 A0A3Q3JJH5 A0A0K0EJZ8 E2A276 A0A195FSL3 A0A3R7SHD2 A0A0P5BIH0 A0A0P5LPU2 A0A1B0CN13 A0A023FJE6 A0A087TQV4 A0A0K0G0S9 A0A3Q3XIY9 A0A0C9R1Z1 A0A0N4Z1T4 A0A3P8VK17 A0A1A9YJW5 A0A1B0BQG0 K9J0G9 A0A158NW19 G3NNI6 B7PR76 B4LQY6 I3ITY5 D3TPF3
A0A194QD47 B0X5D4 A0A1Q3F6Y1 A0A0N8ES10 A0A182GQD9 A0A182TMH6 A0A182WJ45 A0A2M3Z7D0 A0A2M4AL73 A0A2M4AL59 A0A2M4AM84 A0A182PQB0 Q7QJI1 A0A2M3Z7P6 A0A182X3L8 A0A182I3F1 A0A2M4BRR3 A0A084WNF8 A0A2M3Z7U5 A0A182NHR4 A0A182JNC6 W5JJS3 A0A182RNF4 D6W8S4 A0A182M803 A0A182KRH0 A0A2R5L8N2 A0A182VFT1 A0A182QIR8 A0A182FG02 G3MQQ0 A0A182JD95 A0A023GME0 A0A182XZ02 A0A1Y1LXF5 U5ESH2 E9HG12 A0A226CZB8 A0A1E1X607 A0A0P4ZZY0 A0A0L7QJJ9 A0A0P6I319 A0A088ADW9 A0A0P6F5F7 A0A131XLK9 A0A0P6AHU0 L7MB32 A0A224Z7A2 A0A0A9XMT4 A0A023F899 A0A023FX40 A0A0P6EUA5 A0A1J1J880 A0A1B6DYD4 A0A224XEG4 R4G3S1 A0A131Z0J0 A0A1L8E144 A0A0N7Z9G1 T1KER6 A0A3P9I8H2 A0A3P9JWQ4 A0A3B3D2S8 A0A232EVA3 A0A0P5MZL2 A0A0L0CJM0 A0A0P5KRP3 A0A3B3HQ15 A0A0P5VTF6 A0A0T6BBV5 A0A3Q3JJH5 A0A0K0EJZ8 E2A276 A0A195FSL3 A0A3R7SHD2 A0A0P5BIH0 A0A0P5LPU2 A0A1B0CN13 A0A023FJE6 A0A087TQV4 A0A0K0G0S9 A0A3Q3XIY9 A0A0C9R1Z1 A0A0N4Z1T4 A0A3P8VK17 A0A1A9YJW5 A0A1B0BQG0 K9J0G9 A0A158NW19 G3NNI6 B7PR76 B4LQY6 I3ITY5 D3TPF3
Pubmed
26354079
19121390
22118469
26999592
26483478
12364791
+ More
14747013 17210077 24438588 20920257 23761445 18362917 19820115 20966253 22216098 25244985 28004739 21292972 28503490 28049606 25576852 28797301 25401762 26823975 25474469 26830274 27129103 17554307 29451363 28648823 26108605 20798317 24487278 21347285 17994087 25186727 20353571
14747013 17210077 24438588 20920257 23761445 18362917 19820115 20966253 22216098 25244985 28004739 21292972 28503490 28049606 25576852 28797301 25401762 26823975 25474469 26830274 27129103 17554307 29451363 28648823 26108605 20798317 24487278 21347285 17994087 25186727 20353571
EMBL
NWSH01001740
PCG70269.1
ODYU01001772
SOQ38440.1
RSAL01000211
RVE44205.1
+ More
KQ461198 KPJ05950.1 BABH01023463 BABH01023464 AGBW02014068 OWR42324.1 KQ459185 KPJ03344.1 DS232375 EDS40819.1 GFDL01011725 JAV23320.1 GDUN01000770 JAN95149.1 JXUM01015832 KQ560440 KXJ82398.1 GGFM01003663 MBW24414.1 GGFK01008176 MBW41497.1 GGFK01008202 MBW41523.1 GGFK01008560 MBW41881.1 AAAB01008807 EAA04690.2 GGFM01003783 MBW24534.1 APCN01000207 GGFJ01006553 MBW55694.1 ATLV01024599 KE525352 KFB51752.1 GGFM01003757 MBW24508.1 ADMH02001234 ETN63553.1 KQ971312 EEZ98268.2 AXCM01010044 GGLE01001722 MBY05848.1 AXCN02000376 JO844201 AEO35818.1 GBBM01001378 JAC34040.1 GEZM01044492 JAV78179.1 GANO01003244 JAB56627.1 GL732638 EFX69339.1 LNIX01000050 OXA37964.1 GFAC01004697 JAT94491.1 GDIP01209098 GDIP01090962 JAJ14304.1 KQ415080 KOC58822.1 GDIQ01010411 JAN84326.1 GDIQ01061165 JAN33572.1 GEFH01002015 JAP66566.1 GDIP01030452 JAM73263.1 GACK01004675 JAA60359.1 GFPF01011144 MAA22290.1 GBHO01036220 GBHO01036219 GBHO01036217 GBHO01036215 GBHO01036214 GBHO01035577 GBHO01022648 GBHO01022647 GBHO01022646 GBHO01022645 GBHO01022643 GBRD01000858 GBRD01000857 GBRD01000856 GDHC01015302 GDHC01013773 GDHC01013350 JAG07384.1 JAG07385.1 JAG07387.1 JAG07389.1 JAG07390.1 JAG08027.1 JAG20956.1 JAG20957.1 JAG20958.1 JAG20959.1 JAG20961.1 JAG64963.1 JAQ03327.1 JAQ04856.1 JAQ05279.1 GBBI01001220 JAC17492.1 GBBL01001286 JAC26034.1 GDIQ01059013 JAN35724.1 CVRI01000075 CRL08645.1 GEDC01006614 JAS30684.1 GFTR01005569 JAW10857.1 ACPB03007816 GAHY01001975 JAA75535.1 GEDV01003608 JAP84949.1 GFDF01001869 JAV12215.1 GDKW01000643 JAI55952.1 CAEY01000028 NNAY01002022 OXU22286.1 GDIQ01151284 JAL00442.1 JRES01000300 KNC32608.1 GDIQ01180220 GDIQ01062966 JAK71505.1 GDIP01095560 JAM08155.1 LJIG01002147 KRT84793.1 GL435922 EFN72481.1 KQ981280 KYN43431.1 QCYY01004202 ROT61364.1 GDIP01184300 JAJ39102.1 GDIQ01175372 JAK76353.1 AJWK01019616 GBBK01002725 JAC21757.1 KK116354 KFM67493.1 GBYB01000876 GBYB01006055 JAG70643.1 JAG75822.1 JXJN01018610 GABZ01006506 JAA47019.1 ADTU01027694 ADTU01027695 ABJB010271582 DS770988 EEC09098.1 CH940649 EDW64525.2 AERX01031258 AERX01031259 AERX01031260 AERX01031261 EZ423305 ADD19581.1
KQ461198 KPJ05950.1 BABH01023463 BABH01023464 AGBW02014068 OWR42324.1 KQ459185 KPJ03344.1 DS232375 EDS40819.1 GFDL01011725 JAV23320.1 GDUN01000770 JAN95149.1 JXUM01015832 KQ560440 KXJ82398.1 GGFM01003663 MBW24414.1 GGFK01008176 MBW41497.1 GGFK01008202 MBW41523.1 GGFK01008560 MBW41881.1 AAAB01008807 EAA04690.2 GGFM01003783 MBW24534.1 APCN01000207 GGFJ01006553 MBW55694.1 ATLV01024599 KE525352 KFB51752.1 GGFM01003757 MBW24508.1 ADMH02001234 ETN63553.1 KQ971312 EEZ98268.2 AXCM01010044 GGLE01001722 MBY05848.1 AXCN02000376 JO844201 AEO35818.1 GBBM01001378 JAC34040.1 GEZM01044492 JAV78179.1 GANO01003244 JAB56627.1 GL732638 EFX69339.1 LNIX01000050 OXA37964.1 GFAC01004697 JAT94491.1 GDIP01209098 GDIP01090962 JAJ14304.1 KQ415080 KOC58822.1 GDIQ01010411 JAN84326.1 GDIQ01061165 JAN33572.1 GEFH01002015 JAP66566.1 GDIP01030452 JAM73263.1 GACK01004675 JAA60359.1 GFPF01011144 MAA22290.1 GBHO01036220 GBHO01036219 GBHO01036217 GBHO01036215 GBHO01036214 GBHO01035577 GBHO01022648 GBHO01022647 GBHO01022646 GBHO01022645 GBHO01022643 GBRD01000858 GBRD01000857 GBRD01000856 GDHC01015302 GDHC01013773 GDHC01013350 JAG07384.1 JAG07385.1 JAG07387.1 JAG07389.1 JAG07390.1 JAG08027.1 JAG20956.1 JAG20957.1 JAG20958.1 JAG20959.1 JAG20961.1 JAG64963.1 JAQ03327.1 JAQ04856.1 JAQ05279.1 GBBI01001220 JAC17492.1 GBBL01001286 JAC26034.1 GDIQ01059013 JAN35724.1 CVRI01000075 CRL08645.1 GEDC01006614 JAS30684.1 GFTR01005569 JAW10857.1 ACPB03007816 GAHY01001975 JAA75535.1 GEDV01003608 JAP84949.1 GFDF01001869 JAV12215.1 GDKW01000643 JAI55952.1 CAEY01000028 NNAY01002022 OXU22286.1 GDIQ01151284 JAL00442.1 JRES01000300 KNC32608.1 GDIQ01180220 GDIQ01062966 JAK71505.1 GDIP01095560 JAM08155.1 LJIG01002147 KRT84793.1 GL435922 EFN72481.1 KQ981280 KYN43431.1 QCYY01004202 ROT61364.1 GDIP01184300 JAJ39102.1 GDIQ01175372 JAK76353.1 AJWK01019616 GBBK01002725 JAC21757.1 KK116354 KFM67493.1 GBYB01000876 GBYB01006055 JAG70643.1 JAG75822.1 JXJN01018610 GABZ01006506 JAA47019.1 ADTU01027694 ADTU01027695 ABJB010271582 DS770988 EEC09098.1 CH940649 EDW64525.2 AERX01031258 AERX01031259 AERX01031260 AERX01031261 EZ423305 ADD19581.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000005204
UP000007151
UP000053268
+ More
UP000002320 UP000069940 UP000249989 UP000075902 UP000075920 UP000075885 UP000007062 UP000076407 UP000075840 UP000030765 UP000075884 UP000075881 UP000000673 UP000075900 UP000007266 UP000075883 UP000075882 UP000075903 UP000075886 UP000069272 UP000075880 UP000076408 UP000000305 UP000198287 UP000053825 UP000005203 UP000183832 UP000015103 UP000015104 UP000265200 UP000265180 UP000261560 UP000215335 UP000037069 UP000001038 UP000261600 UP000035681 UP000000311 UP000078541 UP000283509 UP000092461 UP000054359 UP000035680 UP000261620 UP000038045 UP000265120 UP000092443 UP000092460 UP000005205 UP000007635 UP000001555 UP000008792 UP000005207
UP000002320 UP000069940 UP000249989 UP000075902 UP000075920 UP000075885 UP000007062 UP000076407 UP000075840 UP000030765 UP000075884 UP000075881 UP000000673 UP000075900 UP000007266 UP000075883 UP000075882 UP000075903 UP000075886 UP000069272 UP000075880 UP000076408 UP000000305 UP000198287 UP000053825 UP000005203 UP000183832 UP000015103 UP000015104 UP000265200 UP000265180 UP000261560 UP000215335 UP000037069 UP000001038 UP000261600 UP000035681 UP000000311 UP000078541 UP000283509 UP000092461 UP000054359 UP000035680 UP000261620 UP000038045 UP000265120 UP000092443 UP000092460 UP000005205 UP000007635 UP000001555 UP000008792 UP000005207
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JEM1
A0A2H1VC69
A0A3S2LUE1
A0A194QLW2
H9JEX9
A0A212ELF7
+ More
A0A194QD47 B0X5D4 A0A1Q3F6Y1 A0A0N8ES10 A0A182GQD9 A0A182TMH6 A0A182WJ45 A0A2M3Z7D0 A0A2M4AL73 A0A2M4AL59 A0A2M4AM84 A0A182PQB0 Q7QJI1 A0A2M3Z7P6 A0A182X3L8 A0A182I3F1 A0A2M4BRR3 A0A084WNF8 A0A2M3Z7U5 A0A182NHR4 A0A182JNC6 W5JJS3 A0A182RNF4 D6W8S4 A0A182M803 A0A182KRH0 A0A2R5L8N2 A0A182VFT1 A0A182QIR8 A0A182FG02 G3MQQ0 A0A182JD95 A0A023GME0 A0A182XZ02 A0A1Y1LXF5 U5ESH2 E9HG12 A0A226CZB8 A0A1E1X607 A0A0P4ZZY0 A0A0L7QJJ9 A0A0P6I319 A0A088ADW9 A0A0P6F5F7 A0A131XLK9 A0A0P6AHU0 L7MB32 A0A224Z7A2 A0A0A9XMT4 A0A023F899 A0A023FX40 A0A0P6EUA5 A0A1J1J880 A0A1B6DYD4 A0A224XEG4 R4G3S1 A0A131Z0J0 A0A1L8E144 A0A0N7Z9G1 T1KER6 A0A3P9I8H2 A0A3P9JWQ4 A0A3B3D2S8 A0A232EVA3 A0A0P5MZL2 A0A0L0CJM0 A0A0P5KRP3 A0A3B3HQ15 A0A0P5VTF6 A0A0T6BBV5 A0A3Q3JJH5 A0A0K0EJZ8 E2A276 A0A195FSL3 A0A3R7SHD2 A0A0P5BIH0 A0A0P5LPU2 A0A1B0CN13 A0A023FJE6 A0A087TQV4 A0A0K0G0S9 A0A3Q3XIY9 A0A0C9R1Z1 A0A0N4Z1T4 A0A3P8VK17 A0A1A9YJW5 A0A1B0BQG0 K9J0G9 A0A158NW19 G3NNI6 B7PR76 B4LQY6 I3ITY5 D3TPF3
A0A194QD47 B0X5D4 A0A1Q3F6Y1 A0A0N8ES10 A0A182GQD9 A0A182TMH6 A0A182WJ45 A0A2M3Z7D0 A0A2M4AL73 A0A2M4AL59 A0A2M4AM84 A0A182PQB0 Q7QJI1 A0A2M3Z7P6 A0A182X3L8 A0A182I3F1 A0A2M4BRR3 A0A084WNF8 A0A2M3Z7U5 A0A182NHR4 A0A182JNC6 W5JJS3 A0A182RNF4 D6W8S4 A0A182M803 A0A182KRH0 A0A2R5L8N2 A0A182VFT1 A0A182QIR8 A0A182FG02 G3MQQ0 A0A182JD95 A0A023GME0 A0A182XZ02 A0A1Y1LXF5 U5ESH2 E9HG12 A0A226CZB8 A0A1E1X607 A0A0P4ZZY0 A0A0L7QJJ9 A0A0P6I319 A0A088ADW9 A0A0P6F5F7 A0A131XLK9 A0A0P6AHU0 L7MB32 A0A224Z7A2 A0A0A9XMT4 A0A023F899 A0A023FX40 A0A0P6EUA5 A0A1J1J880 A0A1B6DYD4 A0A224XEG4 R4G3S1 A0A131Z0J0 A0A1L8E144 A0A0N7Z9G1 T1KER6 A0A3P9I8H2 A0A3P9JWQ4 A0A3B3D2S8 A0A232EVA3 A0A0P5MZL2 A0A0L0CJM0 A0A0P5KRP3 A0A3B3HQ15 A0A0P5VTF6 A0A0T6BBV5 A0A3Q3JJH5 A0A0K0EJZ8 E2A276 A0A195FSL3 A0A3R7SHD2 A0A0P5BIH0 A0A0P5LPU2 A0A1B0CN13 A0A023FJE6 A0A087TQV4 A0A0K0G0S9 A0A3Q3XIY9 A0A0C9R1Z1 A0A0N4Z1T4 A0A3P8VK17 A0A1A9YJW5 A0A1B0BQG0 K9J0G9 A0A158NW19 G3NNI6 B7PR76 B4LQY6 I3ITY5 D3TPF3
PDB
2J9F
E-value=4.84828e-133,
Score=1215
Ontologies
PATHWAY
GO
Topology
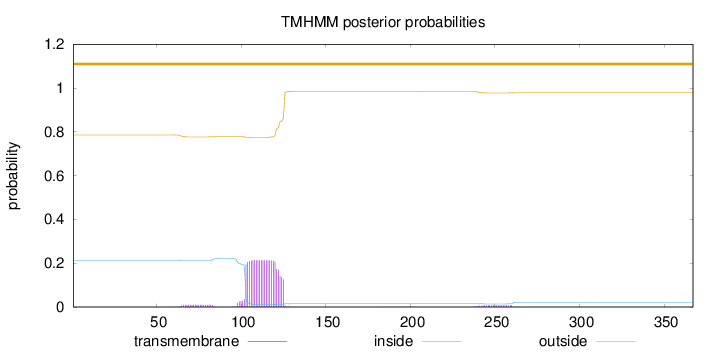
Length:
367
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.05628999999998
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.21407
outside
1 - 367
Population Genetic Test Statistics
Pi
291.353053
Theta
201.828162
Tajima's D
1.615719
CLR
0.098833
CSRT
0.811459427028649
Interpretation
Uncertain
