Gene
KWMTBOMO04924
Pre Gene Modal
BGIBMGA008006
Annotation
PREDICTED:_protein_FAM46C_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.022 Mitochondrial Reliability : 1.06 Nuclear Reliability : 1.272
Sequence
CDS
ATGGACGAGGTGGTGGCCATCCACGGTCGCGGCAACTTCCCTACGCTGCACGTGCGCTTACGCGAGCTTGTAGCTGGGGTGCGAGCCCGCCTGGAGCTGGCGCAGGCGGCTGGCGGGGCCGGGGTCTCCGTACGGGATGTCCGCCTCAACGGCGGCGCTGCCAGCCACGTGCTCGGCGACAACCCACAGCCTTACTCCGACATCGATCTCATCTTCACTGCGGAGCTGCCCACGGCCCGTCACTGCGACCGCGTTAAGGCCGCGGTGCTCGGACACTTGGCCACCCTGCTGCCTCCAGCGACGCCTCGACGTCGCACAACACCTGCTGGACTAAAAGAGGCTTACGTTTCCAAAATGGTCCGCGTGAACTCGGACGGTGACCGCTGGTCTCTCATTTCTCTCGGCAACTCTCGCGGACACAAATCCGTGGAACTCAAGTTCGTGGACAACATGCGCCGTCAGTTCGAGTTCTCGGTAGACTCGTTCCAGATCGCTCTCGACTCCCTGCTCGCGTTCCACGAGTGCGCCCAGCTGCCGATCGGGGAGAACTTTTATCCCACTGTCGTCGGTGAATCCGTTTACGGCGACTTTAACGAGGCCCTGCATCACCTGTCAGAGAAATTGATAGCGACGCGGCAACCCGAAGAGATCCGCGGCGGAGGCCTCCTGAAGTATTGCGCTCTGCTAGCCAAGGGCTACAGAGCCGCGCGTCCCGACAAGATTAAGACCCTCGAACGCTATATGTGCTCGAGGTTCTTCATAGATTTCCCGGAGCTCGGGCAGCAACGCGCCAAGCTCGAGGCTTATTTAAGGAACCATTTCGTGGGCCGAGAAGAGGAAGCGCTCAAGCACAGGTACCTCACGTTGCTGCACAGCGTGGTCCGCGAGTCGACCGTGTGCCTGATGGGCCACGAGCGGCGGCAGACGCTGGCGCTGATCGAGGCGCTGGCGTGCCGGGAACTGTGCGCGCGGACGCCGGTGCTGGTGCAGCCGTACTACCTGTGCGTGTGCACCCCCTGCCCCGCCTGCGCCTACACAGCCTGCTGCGAGTGCTGCCGGCCGGCGCTCTGCCCCGCGTGA
Protein
MDEVVAIHGRGNFPTLHVRLRELVAGVRARLELAQAAGGAGVSVRDVRLNGGAASHVLGDNPQPYSDIDLIFTAELPTARHCDRVKAAVLGHLATLLPPATPRRRTTPAGLKEAYVSKMVRVNSDGDRWSLISLGNSRGHKSVELKFVDNMRRQFEFSVDSFQIALDSLLAFHECAQLPIGENFYPTVVGESVYGDFNEALHHLSEKLIATRQPEEIRGGGLLKYCALLAKGYRAARPDKIKTLERYMCSRFFIDFPELGQQRAKLEAYLRNHFVGREEEALKHRYLTLLHSVVRESTVCLMGHERRQTLALIEALACRELCARTPVLVQPYYLCVCTPCPACAYTACCECCRPALCPA
Summary
Uniprot
H9JEQ9
A0A2A4J146
A0A2H1WZZ0
A0A2A4J2C7
A0A2W1BS97
A0A1E1W2S9
+ More
A0A0L7LJC6 A0A212FFP6 A0A194QK90 A0A194QD07 A0A2J7QHW5 A0A067QXB6 A0A195DIC6 A0A154PD99 A0A0J7L6M9 A0A026WKU3 F4WBR5 A0A158NRG1 A0A151XHQ9 A0A195FSI9 A0A195C3M7 E2AXF4 A0A3L8E499 A0A195BD57 A0A2P8XU67 E9IG33 A0A1B6D1B2 A0A2A3E2W6 E2BC86 A0A0C9R0U4 A0A088ADX0 A0A1Y1LAP4 A0A0L7RGE0 A0A1B6E2G4 A0A310SEL1 A0A2R7WJU7 A0A1B6MHV4 A0A1B6KSE6 A0A1B6LXB9 A0A1W4W7J3 R4WNZ1 A0A0A9XWM8 A0A1B6GD69 A0A1B6HIR8 A0A0K8SXQ4 A0A0A9W2E8 A0A1B6FCN8 A0A023FB06 A0A069DU59 A0A224XR26 A0A1B0DD62 A0A139WMM9 A0A0P4VH25 R4G8S0 A0A1L8DB75 A0A1B0CMQ3 A0A182YFP7 A0A182RA28 A0A1S3DTU1 A0A182WB64 A0A1B6D3Z9 A0A182LX07 A0A182KFS5 U5ETC7 A0NDT1 A0A182HPA6 A0A182PLD5 A0A0K8TR87 A0A182NLZ7 A0A182QJN4 A0A182FLE0 A0A2M3YZ11 A0A2M4BH63 A0A2M3ZFF4 T1PJM0 A0A2M4A152 A0A2M4A196 A0A182IR08 A0A034VHF7 A0A2M4CXY4 A0A2M4CXX8 W8AVT4 A0A1I8N4L2 A0A1J1J9W8 A0A0R3NS11 A0A1I8N4M0 A0A1W4W9U1 A1Z745 A0A0R3NL79 A0A0R1DM50 A0A0Q5WBI7 A0A1W4VYY8 B4QES2 A0A1Q3EXJ9 A0A1Q3EXK7 A0A1Q3EXK2 A0A1Q3EXV4 Q8SXN9 A0A0R3NL15 A0A0R1DLS2
A0A0L7LJC6 A0A212FFP6 A0A194QK90 A0A194QD07 A0A2J7QHW5 A0A067QXB6 A0A195DIC6 A0A154PD99 A0A0J7L6M9 A0A026WKU3 F4WBR5 A0A158NRG1 A0A151XHQ9 A0A195FSI9 A0A195C3M7 E2AXF4 A0A3L8E499 A0A195BD57 A0A2P8XU67 E9IG33 A0A1B6D1B2 A0A2A3E2W6 E2BC86 A0A0C9R0U4 A0A088ADX0 A0A1Y1LAP4 A0A0L7RGE0 A0A1B6E2G4 A0A310SEL1 A0A2R7WJU7 A0A1B6MHV4 A0A1B6KSE6 A0A1B6LXB9 A0A1W4W7J3 R4WNZ1 A0A0A9XWM8 A0A1B6GD69 A0A1B6HIR8 A0A0K8SXQ4 A0A0A9W2E8 A0A1B6FCN8 A0A023FB06 A0A069DU59 A0A224XR26 A0A1B0DD62 A0A139WMM9 A0A0P4VH25 R4G8S0 A0A1L8DB75 A0A1B0CMQ3 A0A182YFP7 A0A182RA28 A0A1S3DTU1 A0A182WB64 A0A1B6D3Z9 A0A182LX07 A0A182KFS5 U5ETC7 A0NDT1 A0A182HPA6 A0A182PLD5 A0A0K8TR87 A0A182NLZ7 A0A182QJN4 A0A182FLE0 A0A2M3YZ11 A0A2M4BH63 A0A2M3ZFF4 T1PJM0 A0A2M4A152 A0A2M4A196 A0A182IR08 A0A034VHF7 A0A2M4CXY4 A0A2M4CXX8 W8AVT4 A0A1I8N4L2 A0A1J1J9W8 A0A0R3NS11 A0A1I8N4M0 A0A1W4W9U1 A1Z745 A0A0R3NL79 A0A0R1DM50 A0A0Q5WBI7 A0A1W4VYY8 B4QES2 A0A1Q3EXJ9 A0A1Q3EXK7 A0A1Q3EXK2 A0A1Q3EXV4 Q8SXN9 A0A0R3NL15 A0A0R1DLS2
Pubmed
19121390
28756777
26227816
22118469
26354079
24845553
+ More
24508170 21719571 21347285 20798317 30249741 29403074 21282665 28004739 23691247 25401762 26823975 25474469 26334808 18362917 19820115 27129103 25244985 12364791 14747013 17210077 26369729 25348373 24495485 25315136 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249
24508170 21719571 21347285 20798317 30249741 29403074 21282665 28004739 23691247 25401762 26823975 25474469 26334808 18362917 19820115 27129103 25244985 12364791 14747013 17210077 26369729 25348373 24495485 25315136 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249
EMBL
BABH01023441
NWSH01003993
PCG65589.1
ODYU01012323
SOQ58577.1
PCG65590.1
+ More
KZ149993 PZC75516.1 GDQN01009774 JAT81280.1 JTDY01000891 KOB75535.1 AGBW02008783 OWR52561.1 KQ461198 KPJ05952.1 KQ459185 KPJ03347.1 NEVH01013964 PNF28180.1 KK852854 KDR15019.1 KQ980824 KYN12591.1 KQ434878 KZC09886.1 LBMM01000446 KMQ98321.1 KK107159 EZA56667.1 GL888066 EGI68343.1 ADTU01023956 KQ982123 KYQ59887.1 KQ981280 KYN43428.1 KQ978317 KYM95205.1 GL443548 EFN61934.1 QOIP01000001 RLU26808.1 KQ976511 KYM82493.1 PYGN01001339 PSN35545.1 GL762910 EFZ20445.1 GEDC01017806 GEDC01006568 JAS19492.1 JAS30730.1 KZ288416 PBC26038.1 GL447256 EFN86713.1 GBYB01000496 JAG70263.1 GEZM01063713 JAV69005.1 KQ414596 KOC69899.1 GEDC01016261 GEDC01005188 GEDC01001334 JAS21037.1 JAS32110.1 JAS35964.1 KQ762129 OAD56174.1 KK854949 PTY19892.1 GEBQ01004495 JAT35482.1 GEBQ01025793 GEBQ01017760 JAT14184.1 JAT22217.1 GEBQ01029197 GEBQ01022522 GEBQ01011629 GEBQ01000265 JAT10780.1 JAT17455.1 JAT28348.1 JAT39712.1 AK417391 BAN20606.1 GBHO01021954 GBRD01007758 GDHC01003112 JAG21650.1 JAG58063.1 JAQ15517.1 GECZ01009384 JAS60385.1 GECU01033106 JAS74600.1 GBRD01007759 GDHC01016752 JAG58062.1 JAQ01877.1 GBHO01041595 JAG02009.1 GECZ01021821 JAS47948.1 GBBI01000599 JAC18113.1 GBGD01001657 JAC87232.1 GFTR01005965 JAW10461.1 AJVK01014352 KQ971312 KYB29116.1 GDKW01002897 JAI53698.1 ACPB03007833 GAHY01000487 JAA77023.1 GFDF01010484 JAV03600.1 AJWK01019040 AJWK01019041 AJWK01019042 AJWK01019043 GEDC01016922 JAS20376.1 AXCM01002767 GANO01002840 JAB57031.1 AAAB01008900 EAU76815.2 APCN01002936 GDAI01000960 JAI16643.1 AXCN02000959 GGFM01000758 MBW21509.1 GGFJ01003236 MBW52377.1 GGFM01006511 MBW27262.1 KA648967 AFP63596.1 GGFK01001225 MBW34546.1 GGFK01001194 MBW34515.1 GAKP01016251 GAKP01016246 JAC42706.1 GGFL01006014 MBW70192.1 GGFL01006008 MBW70186.1 GAMC01013596 GAMC01013595 JAB92960.1 CVRI01000075 CRL08668.1 CM000071 KRT01664.1 AE013599 AAN16139.1 AGB93324.1 KRT01666.1 CM000157 KRJ98316.1 CH954177 KQS70919.1 CM000362 CM002911 EDX06068.1 KMY92084.1 GFDL01015011 JAV20034.1 GFDL01015003 JAV20042.1 GFDL01015006 JAV20039.1 GFDL01015007 JAV20038.1 AY089511 AAF59198.3 AAL90249.1 KRT01665.1 KRJ98315.1
KZ149993 PZC75516.1 GDQN01009774 JAT81280.1 JTDY01000891 KOB75535.1 AGBW02008783 OWR52561.1 KQ461198 KPJ05952.1 KQ459185 KPJ03347.1 NEVH01013964 PNF28180.1 KK852854 KDR15019.1 KQ980824 KYN12591.1 KQ434878 KZC09886.1 LBMM01000446 KMQ98321.1 KK107159 EZA56667.1 GL888066 EGI68343.1 ADTU01023956 KQ982123 KYQ59887.1 KQ981280 KYN43428.1 KQ978317 KYM95205.1 GL443548 EFN61934.1 QOIP01000001 RLU26808.1 KQ976511 KYM82493.1 PYGN01001339 PSN35545.1 GL762910 EFZ20445.1 GEDC01017806 GEDC01006568 JAS19492.1 JAS30730.1 KZ288416 PBC26038.1 GL447256 EFN86713.1 GBYB01000496 JAG70263.1 GEZM01063713 JAV69005.1 KQ414596 KOC69899.1 GEDC01016261 GEDC01005188 GEDC01001334 JAS21037.1 JAS32110.1 JAS35964.1 KQ762129 OAD56174.1 KK854949 PTY19892.1 GEBQ01004495 JAT35482.1 GEBQ01025793 GEBQ01017760 JAT14184.1 JAT22217.1 GEBQ01029197 GEBQ01022522 GEBQ01011629 GEBQ01000265 JAT10780.1 JAT17455.1 JAT28348.1 JAT39712.1 AK417391 BAN20606.1 GBHO01021954 GBRD01007758 GDHC01003112 JAG21650.1 JAG58063.1 JAQ15517.1 GECZ01009384 JAS60385.1 GECU01033106 JAS74600.1 GBRD01007759 GDHC01016752 JAG58062.1 JAQ01877.1 GBHO01041595 JAG02009.1 GECZ01021821 JAS47948.1 GBBI01000599 JAC18113.1 GBGD01001657 JAC87232.1 GFTR01005965 JAW10461.1 AJVK01014352 KQ971312 KYB29116.1 GDKW01002897 JAI53698.1 ACPB03007833 GAHY01000487 JAA77023.1 GFDF01010484 JAV03600.1 AJWK01019040 AJWK01019041 AJWK01019042 AJWK01019043 GEDC01016922 JAS20376.1 AXCM01002767 GANO01002840 JAB57031.1 AAAB01008900 EAU76815.2 APCN01002936 GDAI01000960 JAI16643.1 AXCN02000959 GGFM01000758 MBW21509.1 GGFJ01003236 MBW52377.1 GGFM01006511 MBW27262.1 KA648967 AFP63596.1 GGFK01001225 MBW34546.1 GGFK01001194 MBW34515.1 GAKP01016251 GAKP01016246 JAC42706.1 GGFL01006014 MBW70192.1 GGFL01006008 MBW70186.1 GAMC01013596 GAMC01013595 JAB92960.1 CVRI01000075 CRL08668.1 CM000071 KRT01664.1 AE013599 AAN16139.1 AGB93324.1 KRT01666.1 CM000157 KRJ98316.1 CH954177 KQS70919.1 CM000362 CM002911 EDX06068.1 KMY92084.1 GFDL01015011 JAV20034.1 GFDL01015003 JAV20042.1 GFDL01015006 JAV20039.1 GFDL01015007 JAV20038.1 AY089511 AAF59198.3 AAL90249.1 KRT01665.1 KRJ98315.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000235965 UP000027135 UP000078492 UP000076502 UP000036403 UP000053097 UP000007755 UP000005205 UP000075809 UP000078541 UP000078542 UP000000311 UP000279307 UP000078540 UP000245037 UP000242457 UP000008237 UP000005203 UP000053825 UP000192223 UP000092462 UP000007266 UP000015103 UP000092461 UP000076408 UP000075900 UP000079169 UP000075920 UP000075883 UP000075881 UP000007062 UP000075840 UP000075885 UP000075884 UP000075886 UP000069272 UP000075880 UP000095301 UP000183832 UP000001819 UP000192221 UP000000803 UP000002282 UP000008711 UP000000304
UP000235965 UP000027135 UP000078492 UP000076502 UP000036403 UP000053097 UP000007755 UP000005205 UP000075809 UP000078541 UP000078542 UP000000311 UP000279307 UP000078540 UP000245037 UP000242457 UP000008237 UP000005203 UP000053825 UP000192223 UP000092462 UP000007266 UP000015103 UP000092461 UP000076408 UP000075900 UP000079169 UP000075920 UP000075883 UP000075881 UP000007062 UP000075840 UP000075885 UP000075884 UP000075886 UP000069272 UP000075880 UP000095301 UP000183832 UP000001819 UP000192221 UP000000803 UP000002282 UP000008711 UP000000304
Pfam
PF07984 NTP_transf_7
Interpro
IPR012937
TET5
ProteinModelPortal
H9JEQ9
A0A2A4J146
A0A2H1WZZ0
A0A2A4J2C7
A0A2W1BS97
A0A1E1W2S9
+ More
A0A0L7LJC6 A0A212FFP6 A0A194QK90 A0A194QD07 A0A2J7QHW5 A0A067QXB6 A0A195DIC6 A0A154PD99 A0A0J7L6M9 A0A026WKU3 F4WBR5 A0A158NRG1 A0A151XHQ9 A0A195FSI9 A0A195C3M7 E2AXF4 A0A3L8E499 A0A195BD57 A0A2P8XU67 E9IG33 A0A1B6D1B2 A0A2A3E2W6 E2BC86 A0A0C9R0U4 A0A088ADX0 A0A1Y1LAP4 A0A0L7RGE0 A0A1B6E2G4 A0A310SEL1 A0A2R7WJU7 A0A1B6MHV4 A0A1B6KSE6 A0A1B6LXB9 A0A1W4W7J3 R4WNZ1 A0A0A9XWM8 A0A1B6GD69 A0A1B6HIR8 A0A0K8SXQ4 A0A0A9W2E8 A0A1B6FCN8 A0A023FB06 A0A069DU59 A0A224XR26 A0A1B0DD62 A0A139WMM9 A0A0P4VH25 R4G8S0 A0A1L8DB75 A0A1B0CMQ3 A0A182YFP7 A0A182RA28 A0A1S3DTU1 A0A182WB64 A0A1B6D3Z9 A0A182LX07 A0A182KFS5 U5ETC7 A0NDT1 A0A182HPA6 A0A182PLD5 A0A0K8TR87 A0A182NLZ7 A0A182QJN4 A0A182FLE0 A0A2M3YZ11 A0A2M4BH63 A0A2M3ZFF4 T1PJM0 A0A2M4A152 A0A2M4A196 A0A182IR08 A0A034VHF7 A0A2M4CXY4 A0A2M4CXX8 W8AVT4 A0A1I8N4L2 A0A1J1J9W8 A0A0R3NS11 A0A1I8N4M0 A0A1W4W9U1 A1Z745 A0A0R3NL79 A0A0R1DM50 A0A0Q5WBI7 A0A1W4VYY8 B4QES2 A0A1Q3EXJ9 A0A1Q3EXK7 A0A1Q3EXK2 A0A1Q3EXV4 Q8SXN9 A0A0R3NL15 A0A0R1DLS2
A0A0L7LJC6 A0A212FFP6 A0A194QK90 A0A194QD07 A0A2J7QHW5 A0A067QXB6 A0A195DIC6 A0A154PD99 A0A0J7L6M9 A0A026WKU3 F4WBR5 A0A158NRG1 A0A151XHQ9 A0A195FSI9 A0A195C3M7 E2AXF4 A0A3L8E499 A0A195BD57 A0A2P8XU67 E9IG33 A0A1B6D1B2 A0A2A3E2W6 E2BC86 A0A0C9R0U4 A0A088ADX0 A0A1Y1LAP4 A0A0L7RGE0 A0A1B6E2G4 A0A310SEL1 A0A2R7WJU7 A0A1B6MHV4 A0A1B6KSE6 A0A1B6LXB9 A0A1W4W7J3 R4WNZ1 A0A0A9XWM8 A0A1B6GD69 A0A1B6HIR8 A0A0K8SXQ4 A0A0A9W2E8 A0A1B6FCN8 A0A023FB06 A0A069DU59 A0A224XR26 A0A1B0DD62 A0A139WMM9 A0A0P4VH25 R4G8S0 A0A1L8DB75 A0A1B0CMQ3 A0A182YFP7 A0A182RA28 A0A1S3DTU1 A0A182WB64 A0A1B6D3Z9 A0A182LX07 A0A182KFS5 U5ETC7 A0NDT1 A0A182HPA6 A0A182PLD5 A0A0K8TR87 A0A182NLZ7 A0A182QJN4 A0A182FLE0 A0A2M3YZ11 A0A2M4BH63 A0A2M3ZFF4 T1PJM0 A0A2M4A152 A0A2M4A196 A0A182IR08 A0A034VHF7 A0A2M4CXY4 A0A2M4CXX8 W8AVT4 A0A1I8N4L2 A0A1J1J9W8 A0A0R3NS11 A0A1I8N4M0 A0A1W4W9U1 A1Z745 A0A0R3NL79 A0A0R1DM50 A0A0Q5WBI7 A0A1W4VYY8 B4QES2 A0A1Q3EXJ9 A0A1Q3EXK7 A0A1Q3EXK2 A0A1Q3EXV4 Q8SXN9 A0A0R3NL15 A0A0R1DLS2
Ontologies
PANTHER
Topology
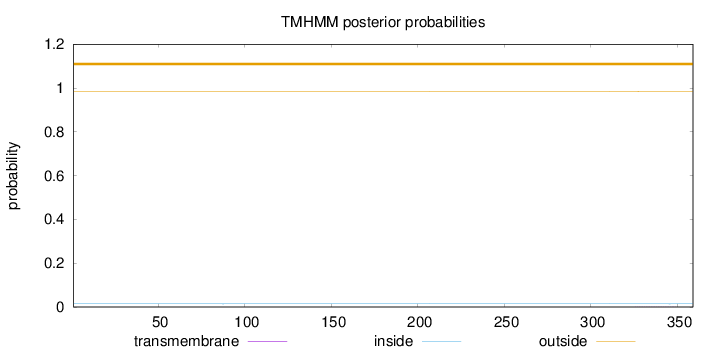
Length:
359
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01774
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.01459
outside
1 - 359
Population Genetic Test Statistics
Pi
197.8719
Theta
161.818791
Tajima's D
0.512922
CLR
0.424405
CSRT
0.51982400879956
Interpretation
Uncertain
