Gene
KWMTBOMO04922
Pre Gene Modal
BGIBMGA008007
Annotation
PREDICTED:_protein_GDAP2_homolog_[Bombyx_mori]
Full name
Protein GDAP2 homolog
Location in the cell
Extracellular Reliability : 1.301 Nuclear Reliability : 1.504
Sequence
CDS
ATGGACAGTTCGGTGTGCGGGATGCGCGCGCCGGGCGAGTGCTGGTCGGTGCCGCGCTGGTCGCAGACGGTGGCCCCTCCCCCCGCCGACTACTCCAACTGCACCCCGCTGCACGCCCAGCCCGTGTTCCTGCACGACGACGCGATCAACGACCGCCTCGCGATATGGCAAGGCGACATAACAAGCTTGAGAGTGGACGCAATTACCAACACCACGGACGAGACGCTGACGGAGAGCAATGAAGTCAGCGACCGCATCATGCAAGTGGCTGGTCCGGATCTGAAAGAAGAGATCATACTCCGGGCACATGAGTGCCGCACCGGCGAGGTGGTGGTGACGCCGGGGTTCGGTCTGCAGTGCCGGCACGTCATCCACACGGTCTGCCCGAAGTACGTGTTGAAGTACCACACCGCGGCCGAGAGCTCGCTGCACAACTGCTACAAGAACGTGCTGCACGCGGCGGGCTCGCTGGGCGCGCGCTCGCTGGCGCTGTGCGTGCTCAACACGCAGCGCCGCAACTTCCCGCCGGACGTCGCCGCGCACGTCGCGCTACGCACGATCCGGCGCTACCTGGAGGGCAACGCACAGCCGCAGCTGGTGGTGGTGGCGGCGGGCGCGGTGGACGCGTCGCTGCTGGGCGCGGTGGCGCCGCTGTACTTCCCGCGGTCGGCGCGCGAGCAGGACGCGGCGCGCTGGCGGCTGCGGGAGGCGGGCGGCCGCGCCGGCGAGCCGCTGCACCACGACCGCCGGATACGCATCATCCACAACCCGCACGAGCACCCGCACCGCGACGTTACAAGAGGTTAA
Protein
MDSSVCGMRAPGECWSVPRWSQTVAPPPADYSNCTPLHAQPVFLHDDAINDRLAIWQGDITSLRVDAITNTTDETLTESNEVSDRIMQVAGPDLKEEIILRAHECRTGEVVVTPGFGLQCRHVIHTVCPKYVLKYHTAAESSLHNCYKNVLHAAGSLGARSLALCVLNTQRRNFPPDVAAHVALRTIRRYLEGNAQPQLVVVAAGAVDASLLGAVAPLYFPRSAREQDAARWRLREAGGRAGEPLHHDRRIRIIHNPHEHPHRDVTRG
Summary
Similarity
Belongs to the GDAP2 family.
Keywords
Complete proteome
Reference proteome
Feature
chain Protein GDAP2 homolog
Uniprot
A0A2A4JIC9
A0A2W1BUT2
A0A2H1WB09
A0A194QK52
A0A194QIS2
A0A1Y1K1D5
+ More
A0A139WML0 A0A139WMS1 E0VU06 A0A2J7PVM8 A0A067R9Y3 A0A1L8DWS6 V5I8E5 A0A182GGM1 A0A182PLD6 A0A1B0GL49 A0A182HPA4 A0A182IP73 A0A182NM75 A0A182X4A1 A0A182LB94 A0A182VM26 A0A182YFP6 Q16HP6 A0A0P6J0P4 A0A1B0AFJ6 A0A182RA27 A0A182WB63 A0A1W4WJA8 A0A1Q3FUS1 Q5TST8 A0A1Q3FUS4 R4WIA7 N6TZF6 A0A1W4WA19 B4P230 A0A2P8XXX4 B3N9U3 B4HR68 A0A0B4KFB9 Q7JUR6 B4QES0 A0A0P4VUH8 A0A3B0JT89 W5J396 A0A0L0C2N8 A0A1I8MME4 B4GDL2 A0A1B6G2K7 A0A1B6JLN4 Q292F9 B3MHT9 A0A2M4DSC5 A0A1I8PBK4 A0A0V0G603 A0A069DVE3 A0A2M4CJ41 A0A0M3QUK0 B4LMJ3 A0A2M4BJI9 A0A023FAK3 A0A224XB52 A0A1B0GCC4 A0A1B6KEQ7 B4J4Y9 A0A084WJH5 T1HPH8 A0A2M4A8T4 A0A2M3YXX2 B4KSX7 A0A2M4A8U5 A0A3M6UPH6 A0A3Q0INM4 A0A146L3B0 A0A151IKD0 B0WFM8 Q16F91 A0A151WU79 F4WXY0 A0A158NYM4 A0A195DE30 A0A0A9YY59 A0A195FTZ9 K7IYW0 A0A0D2X0I0 A0A026X3M8 A0A0L7RGX1 T2M4U9 A0A0J7L113 A0A2A3EBM5 A0A3L8DX81 A0A195B1Q6 A0A232EIQ3 A0A154PEV2 E2BE82 A0A1B0B5X7 V9IBG5 A0A2B4SZS9 E1ZV20
A0A139WML0 A0A139WMS1 E0VU06 A0A2J7PVM8 A0A067R9Y3 A0A1L8DWS6 V5I8E5 A0A182GGM1 A0A182PLD6 A0A1B0GL49 A0A182HPA4 A0A182IP73 A0A182NM75 A0A182X4A1 A0A182LB94 A0A182VM26 A0A182YFP6 Q16HP6 A0A0P6J0P4 A0A1B0AFJ6 A0A182RA27 A0A182WB63 A0A1W4WJA8 A0A1Q3FUS1 Q5TST8 A0A1Q3FUS4 R4WIA7 N6TZF6 A0A1W4WA19 B4P230 A0A2P8XXX4 B3N9U3 B4HR68 A0A0B4KFB9 Q7JUR6 B4QES0 A0A0P4VUH8 A0A3B0JT89 W5J396 A0A0L0C2N8 A0A1I8MME4 B4GDL2 A0A1B6G2K7 A0A1B6JLN4 Q292F9 B3MHT9 A0A2M4DSC5 A0A1I8PBK4 A0A0V0G603 A0A069DVE3 A0A2M4CJ41 A0A0M3QUK0 B4LMJ3 A0A2M4BJI9 A0A023FAK3 A0A224XB52 A0A1B0GCC4 A0A1B6KEQ7 B4J4Y9 A0A084WJH5 T1HPH8 A0A2M4A8T4 A0A2M3YXX2 B4KSX7 A0A2M4A8U5 A0A3M6UPH6 A0A3Q0INM4 A0A146L3B0 A0A151IKD0 B0WFM8 Q16F91 A0A151WU79 F4WXY0 A0A158NYM4 A0A195DE30 A0A0A9YY59 A0A195FTZ9 K7IYW0 A0A0D2X0I0 A0A026X3M8 A0A0L7RGX1 T2M4U9 A0A0J7L113 A0A2A3EBM5 A0A3L8DX81 A0A195B1Q6 A0A232EIQ3 A0A154PEV2 E2BE82 A0A1B0B5X7 V9IBG5 A0A2B4SZS9 E1ZV20
Pubmed
28756777
26354079
28004739
18362917
19820115
20566863
+ More
24845553 26483478 20966253 25244985 17510324 26999592 12364791 14747013 17210077 23691247 23537049 17994087 17550304 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 22936249 27129103 20920257 23761445 26108605 25315136 15632085 18057021 26334808 25474469 24438588 30382153 26823975 21719571 21347285 25401762 20075255 24508170 24065732 30249741 28648823 20798317
24845553 26483478 20966253 25244985 17510324 26999592 12364791 14747013 17210077 23691247 23537049 17994087 17550304 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 22936249 27129103 20920257 23761445 26108605 25315136 15632085 18057021 26334808 25474469 24438588 30382153 26823975 21719571 21347285 25401762 20075255 24508170 24065732 30249741 28648823 20798317
EMBL
NWSH01001292
PCG71817.1
KZ149993
PZC75513.1
ODYU01007453
SOQ50240.1
+ More
KQ461198 KPJ05953.1 KQ459185 KPJ03351.1 GEZM01095920 JAV55263.1 KQ971312 KYB29114.1 KYB29115.1 DS235777 EEB16862.1 NEVH01020956 PNF20360.1 KK852827 KDR15394.1 GFDF01003201 JAV10883.1 GALX01004825 JAB63641.1 JXUM01061863 JXUM01061864 JXUM01061865 JXUM01061866 JXUM01061867 KQ562172 KXJ76502.1 AJWK01034191 AJWK01034192 AJWK01034193 AJWK01034194 AJWK01034195 APCN01002933 CH478152 EAT33775.1 GDUN01000373 JAN95546.1 GFDL01003696 JAV31349.1 AAAB01008900 EAL40460.3 EDO64077.1 GFDL01003695 JAV31350.1 AK417110 BAN20325.1 APGK01048935 APGK01048936 APGK01048937 KB741135 ENN73746.1 CM000157 EDW89231.1 PYGN01001180 PSN36859.1 CH954177 EDV59639.1 CH480816 EDW46811.1 AE013599 AGB93323.1 AY122088 CM000362 CM002911 EDX06066.1 KMY92079.1 GDKW01000394 JAI56201.1 OUUW01000001 SPP74348.1 ADMH02002209 ETN57798.1 JRES01000975 KNC26618.1 CH479181 EDW31669.1 GECZ01013148 JAS56621.1 GECU01007637 GECU01003146 GECU01002227 JAT00070.1 JAT04561.1 JAT05480.1 CM000071 CH902619 EDV36926.1 KPU76529.1 GGFL01016297 MBW80475.1 GECL01003298 JAP02826.1 GBGD01001212 JAC87677.1 GGFL01001145 MBW65323.1 CP012524 ALC40809.1 CH940648 EDW59980.2 GGFJ01004098 MBW53239.1 GBBI01000225 JAC18487.1 GFTR01006790 JAW09636.1 CCAG010007840 GEBQ01030029 JAT09948.1 CH916367 EDW01695.1 ATLV01024033 KE525348 KFB50369.1 ACPB03008271 ACPB03008272 GGFK01003840 MBW37161.1 GGFM01000344 MBW21095.1 CH933808 EDW08474.1 KRG04142.1 GGFK01003717 MBW37038.1 RCHS01001045 RMX55499.1 GDHC01015608 JAQ03021.1 KQ977224 KYN04777.1 DS231919 EDS26334.1 CH478489 EAT32904.1 KQ982748 KYQ51301.1 GL888434 EGI60916.1 ADTU01004071 ADTU01004072 ADTU01004073 KQ980953 KYN11148.1 GBHO01005637 GBRD01012235 JAG37967.1 JAG53589.1 KQ981276 KYN43767.1 KE346360 KJE89094.1 KK107013 EZA62915.1 KQ414596 KOC69936.1 HAAD01000718 CDG66950.1 LBMM01001433 KMQ96326.1 KZ288292 PBC29137.1 QOIP01000003 RLU24902.1 KQ976681 KYM78215.1 NNAY01004185 OXU18225.1 KQ434878 KZC09828.1 GL447768 EFN85927.1 JXJN01008898 JR039001 AEY58478.1 LSMT01000003 PFX34679.1 GL434366 EFN74969.1
KQ461198 KPJ05953.1 KQ459185 KPJ03351.1 GEZM01095920 JAV55263.1 KQ971312 KYB29114.1 KYB29115.1 DS235777 EEB16862.1 NEVH01020956 PNF20360.1 KK852827 KDR15394.1 GFDF01003201 JAV10883.1 GALX01004825 JAB63641.1 JXUM01061863 JXUM01061864 JXUM01061865 JXUM01061866 JXUM01061867 KQ562172 KXJ76502.1 AJWK01034191 AJWK01034192 AJWK01034193 AJWK01034194 AJWK01034195 APCN01002933 CH478152 EAT33775.1 GDUN01000373 JAN95546.1 GFDL01003696 JAV31349.1 AAAB01008900 EAL40460.3 EDO64077.1 GFDL01003695 JAV31350.1 AK417110 BAN20325.1 APGK01048935 APGK01048936 APGK01048937 KB741135 ENN73746.1 CM000157 EDW89231.1 PYGN01001180 PSN36859.1 CH954177 EDV59639.1 CH480816 EDW46811.1 AE013599 AGB93323.1 AY122088 CM000362 CM002911 EDX06066.1 KMY92079.1 GDKW01000394 JAI56201.1 OUUW01000001 SPP74348.1 ADMH02002209 ETN57798.1 JRES01000975 KNC26618.1 CH479181 EDW31669.1 GECZ01013148 JAS56621.1 GECU01007637 GECU01003146 GECU01002227 JAT00070.1 JAT04561.1 JAT05480.1 CM000071 CH902619 EDV36926.1 KPU76529.1 GGFL01016297 MBW80475.1 GECL01003298 JAP02826.1 GBGD01001212 JAC87677.1 GGFL01001145 MBW65323.1 CP012524 ALC40809.1 CH940648 EDW59980.2 GGFJ01004098 MBW53239.1 GBBI01000225 JAC18487.1 GFTR01006790 JAW09636.1 CCAG010007840 GEBQ01030029 JAT09948.1 CH916367 EDW01695.1 ATLV01024033 KE525348 KFB50369.1 ACPB03008271 ACPB03008272 GGFK01003840 MBW37161.1 GGFM01000344 MBW21095.1 CH933808 EDW08474.1 KRG04142.1 GGFK01003717 MBW37038.1 RCHS01001045 RMX55499.1 GDHC01015608 JAQ03021.1 KQ977224 KYN04777.1 DS231919 EDS26334.1 CH478489 EAT32904.1 KQ982748 KYQ51301.1 GL888434 EGI60916.1 ADTU01004071 ADTU01004072 ADTU01004073 KQ980953 KYN11148.1 GBHO01005637 GBRD01012235 JAG37967.1 JAG53589.1 KQ981276 KYN43767.1 KE346360 KJE89094.1 KK107013 EZA62915.1 KQ414596 KOC69936.1 HAAD01000718 CDG66950.1 LBMM01001433 KMQ96326.1 KZ288292 PBC29137.1 QOIP01000003 RLU24902.1 KQ976681 KYM78215.1 NNAY01004185 OXU18225.1 KQ434878 KZC09828.1 GL447768 EFN85927.1 JXJN01008898 JR039001 AEY58478.1 LSMT01000003 PFX34679.1 GL434366 EFN74969.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007266
UP000009046
UP000235965
+ More
UP000027135 UP000069940 UP000249989 UP000075885 UP000092461 UP000075840 UP000075880 UP000075884 UP000076407 UP000075882 UP000075903 UP000076408 UP000008820 UP000092445 UP000075900 UP000075920 UP000192223 UP000007062 UP000019118 UP000192221 UP000002282 UP000245037 UP000008711 UP000001292 UP000000803 UP000000304 UP000268350 UP000000673 UP000037069 UP000095301 UP000008744 UP000001819 UP000007801 UP000095300 UP000092553 UP000008792 UP000092444 UP000001070 UP000030765 UP000015103 UP000009192 UP000275408 UP000079169 UP000078542 UP000002320 UP000075809 UP000007755 UP000005205 UP000078492 UP000078541 UP000002358 UP000008743 UP000053097 UP000053825 UP000036403 UP000242457 UP000279307 UP000078540 UP000215335 UP000076502 UP000008237 UP000092460 UP000225706 UP000000311
UP000027135 UP000069940 UP000249989 UP000075885 UP000092461 UP000075840 UP000075880 UP000075884 UP000076407 UP000075882 UP000075903 UP000076408 UP000008820 UP000092445 UP000075900 UP000075920 UP000192223 UP000007062 UP000019118 UP000192221 UP000002282 UP000245037 UP000008711 UP000001292 UP000000803 UP000000304 UP000268350 UP000000673 UP000037069 UP000095301 UP000008744 UP000001819 UP000007801 UP000095300 UP000092553 UP000008792 UP000092444 UP000001070 UP000030765 UP000015103 UP000009192 UP000275408 UP000079169 UP000078542 UP000002320 UP000075809 UP000007755 UP000005205 UP000078492 UP000078541 UP000002358 UP000008743 UP000053097 UP000053825 UP000036403 UP000242457 UP000279307 UP000078540 UP000215335 UP000076502 UP000008237 UP000092460 UP000225706 UP000000311
PRIDE
Interpro
SUPFAM
SSF52087
SSF52087
Gene 3D
ProteinModelPortal
A0A2A4JIC9
A0A2W1BUT2
A0A2H1WB09
A0A194QK52
A0A194QIS2
A0A1Y1K1D5
+ More
A0A139WML0 A0A139WMS1 E0VU06 A0A2J7PVM8 A0A067R9Y3 A0A1L8DWS6 V5I8E5 A0A182GGM1 A0A182PLD6 A0A1B0GL49 A0A182HPA4 A0A182IP73 A0A182NM75 A0A182X4A1 A0A182LB94 A0A182VM26 A0A182YFP6 Q16HP6 A0A0P6J0P4 A0A1B0AFJ6 A0A182RA27 A0A182WB63 A0A1W4WJA8 A0A1Q3FUS1 Q5TST8 A0A1Q3FUS4 R4WIA7 N6TZF6 A0A1W4WA19 B4P230 A0A2P8XXX4 B3N9U3 B4HR68 A0A0B4KFB9 Q7JUR6 B4QES0 A0A0P4VUH8 A0A3B0JT89 W5J396 A0A0L0C2N8 A0A1I8MME4 B4GDL2 A0A1B6G2K7 A0A1B6JLN4 Q292F9 B3MHT9 A0A2M4DSC5 A0A1I8PBK4 A0A0V0G603 A0A069DVE3 A0A2M4CJ41 A0A0M3QUK0 B4LMJ3 A0A2M4BJI9 A0A023FAK3 A0A224XB52 A0A1B0GCC4 A0A1B6KEQ7 B4J4Y9 A0A084WJH5 T1HPH8 A0A2M4A8T4 A0A2M3YXX2 B4KSX7 A0A2M4A8U5 A0A3M6UPH6 A0A3Q0INM4 A0A146L3B0 A0A151IKD0 B0WFM8 Q16F91 A0A151WU79 F4WXY0 A0A158NYM4 A0A195DE30 A0A0A9YY59 A0A195FTZ9 K7IYW0 A0A0D2X0I0 A0A026X3M8 A0A0L7RGX1 T2M4U9 A0A0J7L113 A0A2A3EBM5 A0A3L8DX81 A0A195B1Q6 A0A232EIQ3 A0A154PEV2 E2BE82 A0A1B0B5X7 V9IBG5 A0A2B4SZS9 E1ZV20
A0A139WML0 A0A139WMS1 E0VU06 A0A2J7PVM8 A0A067R9Y3 A0A1L8DWS6 V5I8E5 A0A182GGM1 A0A182PLD6 A0A1B0GL49 A0A182HPA4 A0A182IP73 A0A182NM75 A0A182X4A1 A0A182LB94 A0A182VM26 A0A182YFP6 Q16HP6 A0A0P6J0P4 A0A1B0AFJ6 A0A182RA27 A0A182WB63 A0A1W4WJA8 A0A1Q3FUS1 Q5TST8 A0A1Q3FUS4 R4WIA7 N6TZF6 A0A1W4WA19 B4P230 A0A2P8XXX4 B3N9U3 B4HR68 A0A0B4KFB9 Q7JUR6 B4QES0 A0A0P4VUH8 A0A3B0JT89 W5J396 A0A0L0C2N8 A0A1I8MME4 B4GDL2 A0A1B6G2K7 A0A1B6JLN4 Q292F9 B3MHT9 A0A2M4DSC5 A0A1I8PBK4 A0A0V0G603 A0A069DVE3 A0A2M4CJ41 A0A0M3QUK0 B4LMJ3 A0A2M4BJI9 A0A023FAK3 A0A224XB52 A0A1B0GCC4 A0A1B6KEQ7 B4J4Y9 A0A084WJH5 T1HPH8 A0A2M4A8T4 A0A2M3YXX2 B4KSX7 A0A2M4A8U5 A0A3M6UPH6 A0A3Q0INM4 A0A146L3B0 A0A151IKD0 B0WFM8 Q16F91 A0A151WU79 F4WXY0 A0A158NYM4 A0A195DE30 A0A0A9YY59 A0A195FTZ9 K7IYW0 A0A0D2X0I0 A0A026X3M8 A0A0L7RGX1 T2M4U9 A0A0J7L113 A0A2A3EBM5 A0A3L8DX81 A0A195B1Q6 A0A232EIQ3 A0A154PEV2 E2BE82 A0A1B0B5X7 V9IBG5 A0A2B4SZS9 E1ZV20
PDB
4UML
E-value=1.51098e-40,
Score=416
Ontologies
Topology
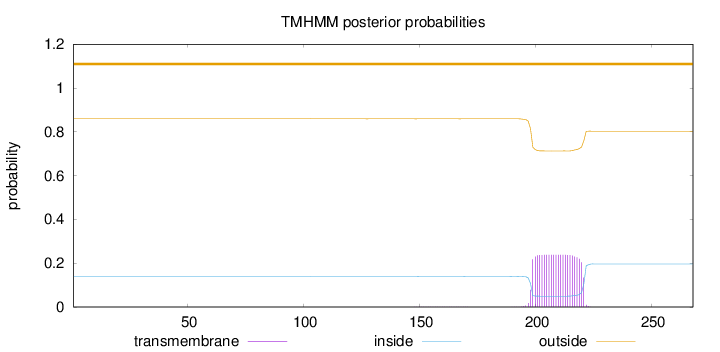
Length:
268
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.40356
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14060
outside
1 - 268
Population Genetic Test Statistics
Pi
154.662224
Theta
165.42756
Tajima's D
1.41437
CLR
185.284462
CSRT
0.766261686915654
Interpretation
Uncertain
