Pre Gene Modal
BGIBMGA008009
Annotation
PREDICTED:_protein_GDAP2_homolog_isoform_X2_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.5 PlasmaMembrane Reliability : 1.24
Sequence
CDS
ATGGCGATGGACACTATGGCAGTGGAATTGCCAGATTTAACCACAACTGCTCCTCCACCAGCCGCAGAAGAAACAGCCGAAGAAAAATATGAAAGACTGCTGCGGCGTGCCAAGTCCGAAGACCTGAGCGAGGTATCTGGGATCGGATGTCTATATCAGAGCGGCGTGGATCGACTGGGTCGGCCCGTGGTCGTCTTCATCGGGAAATGGTTCCCCATTGGGGACATAGACTTGGAAAAGGCACTCCTATACCTGATCAAACTCCTGGATCCGATCGTGCGCGGTGACTACGTGATCGCGTACTTCCACACGTTGGCCTCGTCGGCGAACCATCCGCCCTTTTCGTGGCTCAAGGAGGTGTACACGGTGCTGCCTTACAAATACAAGAAGAATCTGAAAGCTTTCTACATTGTTCATCCTACGTTCTGGACTAAGATGATGACGTGGTGGTTCACGACGTTCATGGCGCCGGCGATAAAGGCCAAGGTCCACAGTCTGCCCGGCGTCGAGTACCTGTACTCCGTCATGCCGCGCGACCAGCTCGAGGTGCCGGCCTTCGTCACGGAGTACGACATGACGATAAACGGTCTCCACTATTTCCAGCCGGACACGACTAGTACAGTCCTCTGCGAACATTTGTCCCTTCAGTACGACTGA
Protein
MAMDTMAVELPDLTTTAPPPAAEETAEEKYERLLRRAKSEDLSEVSGIGCLYQSGVDRLGRPVVVFIGKWFPIGDIDLEKALLYLIKLLDPIVRGDYVIAYFHTLASSANHPPFSWLKEVYTVLPYKYKKNLKAFYIVHPTFWTKMMTWWFTTFMAPAIKAKVHSLPGVEYLYSVMPRDQLEVPAFVTEYDMTINGLHYFQPDTTSTVLCEHLSLQYD
Summary
Uniprot
A0A194QCT5
A0A2H1WA41
A0A2A4JFC3
A0A194QLW7
A0A2W1BKA9
A0A1E1WQW5
+ More
H9JER2 V5GPW7 A0A1L8DNB5 A0A1L8DN29 Q16U54 T1DEY3 U5EMK6 A7UT51 A0A182VH17 A0A212F2Y8 A0A182NM74 V5I8E5 A0A2M4CU60 A0A2M3YXZ3 A0A182FLE2 A0A182UFW3 A0A1Y1K1C8 A0A1B6DD50 A0A182MRB7 A0A2R7W9B1 A0A182KE55 A0A182WB62 A0A182X4A0 A0A182QP13 A0A182PLD7 A0A182HPA3 A7UT54 A0A182JBZ2 A0A2M3YY16 A0A139WML0 W8AV21 A0A1Q3EUD5 A0A139WMS1 A0A1B6DRA1 A0A1B6CID3 A0A1B6IJ59 A0A0B4KEL9 A0A034VL93 B0XCG5 A0A0P9AF70 W5J4R2 A0A034VL91 A0A1W4VZ92 A0A0P8XPM1 A0A1I8NZD3 A0A0K8V3U3 A0A0A1WWK7 A0A1L8DL02 A0A0J9TVL4 A1Z742 A1Z741 A0A0R1DTA0 A0A1W4WBF8 A0A0J9R7N8 C9QPK4 A0A0A1XLV9 A0A1B6C681 A0A182RA26 T1PA85 A0A0R3NLB4 T1DSB1 A0A1W4VZ85 A0A0R1DM33 A0A0Q5WQ38 D1Z396 A0A0J9R7F9 V9IDS7 A0A0R3NS55 A0A0Q9X766 A0A2J7PVM8 B0WFM7 A0A1B6L219 A0A1A9WJZ3 A0A0Q9X7R8 A0A1A9YN14 A0A1A9V1Z1 A0A1B0B5X4 A0A1L8DWS6 A0A2M4CJ41 A0A2M4A8U5 A0A0A9Z837 A0A1Y1K1D5 A0A2M4BJI9 A0A2M4A8T4 A0A2M3YXX2 R4WIA7 W8BUD7 A0A182LB93 Q5TST8 A0A195DE30 A0A158NYM4 A0A1W4WJA8 A0A195FTZ9 A0A0A9Z0I5
H9JER2 V5GPW7 A0A1L8DNB5 A0A1L8DN29 Q16U54 T1DEY3 U5EMK6 A7UT51 A0A182VH17 A0A212F2Y8 A0A182NM74 V5I8E5 A0A2M4CU60 A0A2M3YXZ3 A0A182FLE2 A0A182UFW3 A0A1Y1K1C8 A0A1B6DD50 A0A182MRB7 A0A2R7W9B1 A0A182KE55 A0A182WB62 A0A182X4A0 A0A182QP13 A0A182PLD7 A0A182HPA3 A7UT54 A0A182JBZ2 A0A2M3YY16 A0A139WML0 W8AV21 A0A1Q3EUD5 A0A139WMS1 A0A1B6DRA1 A0A1B6CID3 A0A1B6IJ59 A0A0B4KEL9 A0A034VL93 B0XCG5 A0A0P9AF70 W5J4R2 A0A034VL91 A0A1W4VZ92 A0A0P8XPM1 A0A1I8NZD3 A0A0K8V3U3 A0A0A1WWK7 A0A1L8DL02 A0A0J9TVL4 A1Z742 A1Z741 A0A0R1DTA0 A0A1W4WBF8 A0A0J9R7N8 C9QPK4 A0A0A1XLV9 A0A1B6C681 A0A182RA26 T1PA85 A0A0R3NLB4 T1DSB1 A0A1W4VZ85 A0A0R1DM33 A0A0Q5WQ38 D1Z396 A0A0J9R7F9 V9IDS7 A0A0R3NS55 A0A0Q9X766 A0A2J7PVM8 B0WFM7 A0A1B6L219 A0A1A9WJZ3 A0A0Q9X7R8 A0A1A9YN14 A0A1A9V1Z1 A0A1B0B5X4 A0A1L8DWS6 A0A2M4CJ41 A0A2M4A8U5 A0A0A9Z837 A0A1Y1K1D5 A0A2M4BJI9 A0A2M4A8T4 A0A2M3YXX2 R4WIA7 W8BUD7 A0A182LB93 Q5TST8 A0A195DE30 A0A158NYM4 A0A1W4WJA8 A0A195FTZ9 A0A0A9Z0I5
Pubmed
26354079
28756777
19121390
17510324
24330624
12364791
+ More
14747013 17210077 22118469 28004739 18362917 19820115 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25348373 17994087 20920257 23761445 25830018 22936249 26109357 26109356 17550304 15632085 25401762 23691247 20966253 21347285
14747013 17210077 22118469 28004739 18362917 19820115 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25348373 17994087 20920257 23761445 25830018 22936249 26109357 26109356 17550304 15632085 25401762 23691247 20966253 21347285
EMBL
KQ459185
KPJ03353.1
ODYU01007169
SOQ49712.1
NWSH01001738
PCG70274.1
+ More
KQ461198 KPJ05955.1 KZ149993 PZC75512.1 GDQN01002621 GDQN01001655 JAT88433.1 JAT89399.1 BABH01023427 BABH01023428 GALX01004824 JAB63642.1 GFDF01006217 JAV07867.1 GFDF01006216 JAV07868.1 CH477631 EAT38079.1 GALA01000867 JAA93985.1 GANO01004443 JAB55428.1 AAAB01008900 EDO64079.1 EDO64080.1 AGBW02010653 OWR48099.1 GALX01004825 JAB63641.1 GGFL01004200 MBW68378.1 GGFM01000388 MBW21139.1 GEZM01095921 JAV55262.1 GEDC01013700 JAS23598.1 AXCM01003682 AXCM01003683 KK854411 PTY15630.1 AXCN02000946 APCN01002933 EDO64078.1 GGFM01000394 MBW21145.1 KQ971312 KYB29114.1 GAMC01013815 GAMC01013814 JAB92740.1 GFDL01016115 JAV18930.1 KYB29115.1 GEDC01009165 JAS28133.1 GEDC01024203 JAS13095.1 GECU01020749 JAS86957.1 AE013599 AGB93322.1 GAKP01015743 JAC43209.1 DS232691 EDS44795.1 CH902619 KPU76528.1 ADMH02002209 ETN57799.1 GAKP01015748 GAKP01015744 JAC43204.1 KPU76530.1 GDHF01018778 JAI33536.1 GBXI01010848 JAD03444.1 GFDF01006946 JAV07138.1 CM002911 KMY92080.1 BT133240 AAG22307.1 AFB18663.1 AAG22306.1 CM000157 KRJ98313.1 KMY92081.1 BT100118 ACX83598.1 GBXI01002345 JAD11947.1 GEDC01028286 JAS09012.1 KA645647 AFP60276.1 CM000071 KRT01668.1 GAMD01000255 JAB01336.1 KRJ98314.1 CH954177 KQS70920.1 BT120068 ADA69465.1 AFH07946.1 KMY92082.1 JR039002 AEY58479.1 KRT01667.1 CH933808 KRG04143.1 NEVH01020956 PNF20360.1 DS231919 EDS26333.1 GEBQ01022215 JAT17762.1 KRG04141.1 JXJN01008899 GFDF01003201 JAV10883.1 GGFL01001145 MBW65323.1 GGFK01003717 MBW37038.1 GBHO01005634 GBRD01012234 JAG37970.1 JAG53590.1 GEZM01095920 JAV55263.1 GGFJ01004098 MBW53239.1 GGFK01003840 MBW37161.1 GGFM01000344 MBW21095.1 AK417110 BAN20325.1 GAMC01013818 GAMC01013817 GAMC01013816 JAB92739.1 EAL40460.3 EDO64077.1 KQ980953 KYN11148.1 ADTU01004071 ADTU01004072 ADTU01004073 KQ981276 KYN43767.1 GBHO01005640 JAG37964.1
KQ461198 KPJ05955.1 KZ149993 PZC75512.1 GDQN01002621 GDQN01001655 JAT88433.1 JAT89399.1 BABH01023427 BABH01023428 GALX01004824 JAB63642.1 GFDF01006217 JAV07867.1 GFDF01006216 JAV07868.1 CH477631 EAT38079.1 GALA01000867 JAA93985.1 GANO01004443 JAB55428.1 AAAB01008900 EDO64079.1 EDO64080.1 AGBW02010653 OWR48099.1 GALX01004825 JAB63641.1 GGFL01004200 MBW68378.1 GGFM01000388 MBW21139.1 GEZM01095921 JAV55262.1 GEDC01013700 JAS23598.1 AXCM01003682 AXCM01003683 KK854411 PTY15630.1 AXCN02000946 APCN01002933 EDO64078.1 GGFM01000394 MBW21145.1 KQ971312 KYB29114.1 GAMC01013815 GAMC01013814 JAB92740.1 GFDL01016115 JAV18930.1 KYB29115.1 GEDC01009165 JAS28133.1 GEDC01024203 JAS13095.1 GECU01020749 JAS86957.1 AE013599 AGB93322.1 GAKP01015743 JAC43209.1 DS232691 EDS44795.1 CH902619 KPU76528.1 ADMH02002209 ETN57799.1 GAKP01015748 GAKP01015744 JAC43204.1 KPU76530.1 GDHF01018778 JAI33536.1 GBXI01010848 JAD03444.1 GFDF01006946 JAV07138.1 CM002911 KMY92080.1 BT133240 AAG22307.1 AFB18663.1 AAG22306.1 CM000157 KRJ98313.1 KMY92081.1 BT100118 ACX83598.1 GBXI01002345 JAD11947.1 GEDC01028286 JAS09012.1 KA645647 AFP60276.1 CM000071 KRT01668.1 GAMD01000255 JAB01336.1 KRJ98314.1 CH954177 KQS70920.1 BT120068 ADA69465.1 AFH07946.1 KMY92082.1 JR039002 AEY58479.1 KRT01667.1 CH933808 KRG04143.1 NEVH01020956 PNF20360.1 DS231919 EDS26333.1 GEBQ01022215 JAT17762.1 KRG04141.1 JXJN01008899 GFDF01003201 JAV10883.1 GGFL01001145 MBW65323.1 GGFK01003717 MBW37038.1 GBHO01005634 GBRD01012234 JAG37970.1 JAG53590.1 GEZM01095920 JAV55263.1 GGFJ01004098 MBW53239.1 GGFK01003840 MBW37161.1 GGFM01000344 MBW21095.1 AK417110 BAN20325.1 GAMC01013818 GAMC01013817 GAMC01013816 JAB92739.1 EAL40460.3 EDO64077.1 KQ980953 KYN11148.1 ADTU01004071 ADTU01004072 ADTU01004073 KQ981276 KYN43767.1 GBHO01005640 JAG37964.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000005204
UP000008820
UP000007062
+ More
UP000075903 UP000007151 UP000075884 UP000069272 UP000075902 UP000075883 UP000075881 UP000075920 UP000076407 UP000075886 UP000075885 UP000075840 UP000075880 UP000007266 UP000000803 UP000002320 UP000007801 UP000000673 UP000192221 UP000095300 UP000002282 UP000075900 UP000001819 UP000008711 UP000009192 UP000235965 UP000091820 UP000092443 UP000078200 UP000092460 UP000075882 UP000078492 UP000005205 UP000192223 UP000078541
UP000075903 UP000007151 UP000075884 UP000069272 UP000075902 UP000075883 UP000075881 UP000075920 UP000076407 UP000075886 UP000075885 UP000075840 UP000075880 UP000007266 UP000000803 UP000002320 UP000007801 UP000000673 UP000192221 UP000095300 UP000002282 UP000075900 UP000001819 UP000008711 UP000009192 UP000235965 UP000091820 UP000092443 UP000078200 UP000092460 UP000075882 UP000078492 UP000005205 UP000192223 UP000078541
Interpro
SUPFAM
SSF52087
SSF52087
Gene 3D
ProteinModelPortal
A0A194QCT5
A0A2H1WA41
A0A2A4JFC3
A0A194QLW7
A0A2W1BKA9
A0A1E1WQW5
+ More
H9JER2 V5GPW7 A0A1L8DNB5 A0A1L8DN29 Q16U54 T1DEY3 U5EMK6 A7UT51 A0A182VH17 A0A212F2Y8 A0A182NM74 V5I8E5 A0A2M4CU60 A0A2M3YXZ3 A0A182FLE2 A0A182UFW3 A0A1Y1K1C8 A0A1B6DD50 A0A182MRB7 A0A2R7W9B1 A0A182KE55 A0A182WB62 A0A182X4A0 A0A182QP13 A0A182PLD7 A0A182HPA3 A7UT54 A0A182JBZ2 A0A2M3YY16 A0A139WML0 W8AV21 A0A1Q3EUD5 A0A139WMS1 A0A1B6DRA1 A0A1B6CID3 A0A1B6IJ59 A0A0B4KEL9 A0A034VL93 B0XCG5 A0A0P9AF70 W5J4R2 A0A034VL91 A0A1W4VZ92 A0A0P8XPM1 A0A1I8NZD3 A0A0K8V3U3 A0A0A1WWK7 A0A1L8DL02 A0A0J9TVL4 A1Z742 A1Z741 A0A0R1DTA0 A0A1W4WBF8 A0A0J9R7N8 C9QPK4 A0A0A1XLV9 A0A1B6C681 A0A182RA26 T1PA85 A0A0R3NLB4 T1DSB1 A0A1W4VZ85 A0A0R1DM33 A0A0Q5WQ38 D1Z396 A0A0J9R7F9 V9IDS7 A0A0R3NS55 A0A0Q9X766 A0A2J7PVM8 B0WFM7 A0A1B6L219 A0A1A9WJZ3 A0A0Q9X7R8 A0A1A9YN14 A0A1A9V1Z1 A0A1B0B5X4 A0A1L8DWS6 A0A2M4CJ41 A0A2M4A8U5 A0A0A9Z837 A0A1Y1K1D5 A0A2M4BJI9 A0A2M4A8T4 A0A2M3YXX2 R4WIA7 W8BUD7 A0A182LB93 Q5TST8 A0A195DE30 A0A158NYM4 A0A1W4WJA8 A0A195FTZ9 A0A0A9Z0I5
H9JER2 V5GPW7 A0A1L8DNB5 A0A1L8DN29 Q16U54 T1DEY3 U5EMK6 A7UT51 A0A182VH17 A0A212F2Y8 A0A182NM74 V5I8E5 A0A2M4CU60 A0A2M3YXZ3 A0A182FLE2 A0A182UFW3 A0A1Y1K1C8 A0A1B6DD50 A0A182MRB7 A0A2R7W9B1 A0A182KE55 A0A182WB62 A0A182X4A0 A0A182QP13 A0A182PLD7 A0A182HPA3 A7UT54 A0A182JBZ2 A0A2M3YY16 A0A139WML0 W8AV21 A0A1Q3EUD5 A0A139WMS1 A0A1B6DRA1 A0A1B6CID3 A0A1B6IJ59 A0A0B4KEL9 A0A034VL93 B0XCG5 A0A0P9AF70 W5J4R2 A0A034VL91 A0A1W4VZ92 A0A0P8XPM1 A0A1I8NZD3 A0A0K8V3U3 A0A0A1WWK7 A0A1L8DL02 A0A0J9TVL4 A1Z742 A1Z741 A0A0R1DTA0 A0A1W4WBF8 A0A0J9R7N8 C9QPK4 A0A0A1XLV9 A0A1B6C681 A0A182RA26 T1PA85 A0A0R3NLB4 T1DSB1 A0A1W4VZ85 A0A0R1DM33 A0A0Q5WQ38 D1Z396 A0A0J9R7F9 V9IDS7 A0A0R3NS55 A0A0Q9X766 A0A2J7PVM8 B0WFM7 A0A1B6L219 A0A1A9WJZ3 A0A0Q9X7R8 A0A1A9YN14 A0A1A9V1Z1 A0A1B0B5X4 A0A1L8DWS6 A0A2M4CJ41 A0A2M4A8U5 A0A0A9Z837 A0A1Y1K1D5 A0A2M4BJI9 A0A2M4A8T4 A0A2M3YXX2 R4WIA7 W8BUD7 A0A182LB93 Q5TST8 A0A195DE30 A0A158NYM4 A0A1W4WJA8 A0A195FTZ9 A0A0A9Z0I5
PDB
6R3V
E-value=1.98275e-10,
Score=155
Ontologies
GO
Topology
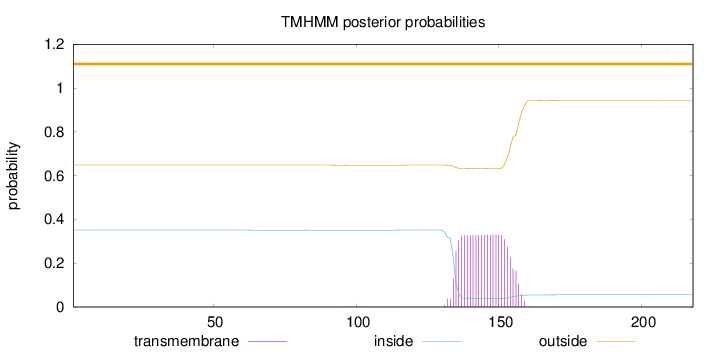
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.13142
Exp number, first 60 AAs:
0.00045
Total prob of N-in:
0.35123
outside
1 - 218
Population Genetic Test Statistics
Pi
207.259516
Theta
175.538913
Tajima's D
1.238773
CLR
0.375096
CSRT
0.722713864306785
Interpretation
Uncertain
