Gene
KWMTBOMO04916
Pre Gene Modal
BGIBMGA008073
Annotation
hypothetical_protein_KGM_01342_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.153 Nuclear Reliability : 1.006
Sequence
CDS
ATGCCCGTGATGCCCAGGGACATCCCGGTGGACATGAAGTACATCGCGGACCCGACGATGCACTCCCTGCCGGCCGTCACCGCCGCGCCCAACGGCAAGTGGCTCGCGTGCCAGTCCATGGACAACAAGGTCATCGTGTTCAGCGCGCTCAACCGCTTCAAGATGAACAGGAAGAAGACGTTCACCGGGCACATGGTGGCCGGGTACGCCTGCACCGTCGACTTCTCCCCGGACATGAGGTAA
Protein
MPVMPRDIPVDMKYIADPTMHSLPAVTAAPNGKWLACQSMDNKVIVFSALNRFKMNRKKTFTGHMVAGYACTVDFSPDMR
Summary
Uniprot
H9JEX6
A0A2J7R200
A0A1B0DFE9
A0A224XDZ3
A0A023F4L3
A0A232FIR0
+ More
K7J197 N6TSG8 U4TXH7 A0A1L8DG53 A0A195CJ25 A0A310SQC6 A0A1J1J2U2 A0A2R7X4G2 A0A1A9XN20 A0A0L7RDQ3 A0A1I8NU20 E0VQD0 A0A1A9ZZP9 A0A154PB20 A0A1B0FRH7 A0A3L8DQ43 A0A195F2I9 A0A158NFU1 A0A195E6W1 A0A026WYQ6 F4X6R4 A0A151XC95 A0A0J7KS73 A0A195B2R1 E2ACA9 A0A0P4WTK3 A0A0P5KBL1 A0A2P6L0K4 A0A226D9P0 A0A162EE21 A0A194QK57 A0A194QIS7 V5GIC3 A0A0P5G7C8 A0A2G9RUK5 A0A2A4JPL1 A0A0B6ZXN2 A0A069DYH3 A0A087UVE6 K1QLM6 A0A1W4X2G2 A0A1Y1N8F7 A0A2R9CDQ9 K9KCK8 A0A1E1WKN5 A0A2R9CDR7 A0A2I3TNU2 G8F2I8 A0A0G2JW64 A0A094L6E0 A0A2C9K688 A0A093QN90 A0A2P4T741 A0A091SQY4 A0A3Q3SUV7 A0A093IMA9 T1JN96 D6X3W5 A0A0T6AUB0 A0A034VRU3 A0A0V0GCK5 A0A3Q1EQA8 A0A2I2V1E3 A0A3S1AGN9 F7CR49 A0A3Q1B0G7 A0A3P8S795 A0A3B5A626 A0A2K5Z5N2 A0A2I3H9S3 A0A067RLH9 A0A341B3G8 A0A226NTM2 A0A2I3T8T6 Q5SRN1 M1EIH2 A0A093DWF8 A0A2K6BF43 A0A2K5TZC3 A0A2K5Z5S4 A0A096NFM5 A0A2K5KF60 A0A182T3I1 A0A2K6JP06 A0A2K6P181 A0A2K5C2C0 H2PK17 A0A2J8U1H4 A0A2K5PDT0 M7CBG4 A0A182QRA8 A0A2I3MLP9 A0A182K1F3 U3K871
K7J197 N6TSG8 U4TXH7 A0A1L8DG53 A0A195CJ25 A0A310SQC6 A0A1J1J2U2 A0A2R7X4G2 A0A1A9XN20 A0A0L7RDQ3 A0A1I8NU20 E0VQD0 A0A1A9ZZP9 A0A154PB20 A0A1B0FRH7 A0A3L8DQ43 A0A195F2I9 A0A158NFU1 A0A195E6W1 A0A026WYQ6 F4X6R4 A0A151XC95 A0A0J7KS73 A0A195B2R1 E2ACA9 A0A0P4WTK3 A0A0P5KBL1 A0A2P6L0K4 A0A226D9P0 A0A162EE21 A0A194QK57 A0A194QIS7 V5GIC3 A0A0P5G7C8 A0A2G9RUK5 A0A2A4JPL1 A0A0B6ZXN2 A0A069DYH3 A0A087UVE6 K1QLM6 A0A1W4X2G2 A0A1Y1N8F7 A0A2R9CDQ9 K9KCK8 A0A1E1WKN5 A0A2R9CDR7 A0A2I3TNU2 G8F2I8 A0A0G2JW64 A0A094L6E0 A0A2C9K688 A0A093QN90 A0A2P4T741 A0A091SQY4 A0A3Q3SUV7 A0A093IMA9 T1JN96 D6X3W5 A0A0T6AUB0 A0A034VRU3 A0A0V0GCK5 A0A3Q1EQA8 A0A2I2V1E3 A0A3S1AGN9 F7CR49 A0A3Q1B0G7 A0A3P8S795 A0A3B5A626 A0A2K5Z5N2 A0A2I3H9S3 A0A067RLH9 A0A341B3G8 A0A226NTM2 A0A2I3T8T6 Q5SRN1 M1EIH2 A0A093DWF8 A0A2K6BF43 A0A2K5TZC3 A0A2K5Z5S4 A0A096NFM5 A0A2K5KF60 A0A182T3I1 A0A2K6JP06 A0A2K6P181 A0A2K5C2C0 H2PK17 A0A2J8U1H4 A0A2K5PDT0 M7CBG4 A0A182QRA8 A0A2I3MLP9 A0A182K1F3 U3K871
Pubmed
19121390
25474469
28648823
20075255
23537049
20566863
+ More
30249741 21347285 24508170 21719571 20798317 26354079 26334808 22992520 28004739 22722832 16136131 22002653 15057822 15562597 18362917 19820115 25348373 17975172 24845553 24621616 14574404 18669648 19690332 20068231 21406692 23186163 24275569 23236062 25362486 23624526
30249741 21347285 24508170 21719571 20798317 26354079 26334808 22992520 28004739 22722832 16136131 22002653 15057822 15562597 18362917 19820115 25348373 17975172 24845553 24621616 14574404 18669648 19690332 20068231 21406692 23186163 24275569 23236062 25362486 23624526
EMBL
BABH01023427
NEVH01008202
PNF34866.1
AJVK01058702
GFTR01007222
JAW09204.1
+ More
GBBI01002465 JAC16247.1 NNAY01000140 OXU30626.1 APGK01057068 KB741277 ENN71276.1 KB631617 ERL84703.1 GFDF01008770 JAV05314.1 KQ977754 KYN00069.1 KQ761768 OAD57125.1 CVRI01000067 CRL06687.1 KK856889 PTY26531.1 KQ414613 KOC68925.1 DS235418 EEB15586.1 KQ434864 KZC09031.1 CCAG010008016 QOIP01000005 RLU22353.1 KQ981864 KYN34304.1 ADTU01014740 KQ979568 KYN20831.1 KK107063 EZA61175.1 GL888818 EGI57846.1 KQ982314 KYQ57967.1 LBMM01003834 KMQ93069.1 KQ976662 KYM78489.1 GL438491 EFN68899.1 GDRN01007567 JAI67946.1 GDIQ01186460 JAK65265.1 MWRG01002896 PRD32117.1 LNIX01000028 OXA41860.1 LRGB01002011 KZS09650.1 KQ461198 KPJ05958.1 KQ459185 KPJ03356.1 GALX01007084 JAB61382.1 GDIQ01246234 JAK05491.1 KV931317 PIO31589.1 NWSH01000969 PCG73352.1 HACG01025751 CEK72616.1 GBGD01002350 JAC86539.1 KK121841 KFM81335.1 JH817213 EKC34793.1 GEZM01010125 JAV94181.1 AJFE02016426 AJFE02016427 AJFE02016428 AJFE02016429 JL621568 AEP99484.1 GDQN01003628 JAT87426.1 AACZ04054326 JH330059 EHH61502.1 AABR07045469 AABR07045470 AABR07045471 AABR07045472 KL278156 KFZ66933.1 KL419269 KFW87855.1 PPHD01006509 POI32166.1 KK483166 KFQ61127.1 KK592048 KFW00762.1 JH431704 KQ971379 EEZ97476.1 LJIG01022834 KRT78475.1 GAKP01014699 JAC44253.1 GECL01000528 JAP05596.1 AANG04003966 RQTK01000009 RUS91544.1 ADFV01164853 ADFV01164854 ADFV01164855 ADFV01164856 KK852438 KDR23898.1 AWGT02001832 OXB70419.1 NBAG03000210 PNI87629.1 AL445068 AL671518 JP007299 AER95896.1 KL232255 KFV06501.1 AQIA01049409 AHZZ02025974 ABGA01320989 ABGA01320990 ABGA01320991 ABGA01320992 ABGA01320993 NDHI03003476 PNJ39113.1 KB520915 EMP38047.1 AXCN02002066 AGTO01010121
GBBI01002465 JAC16247.1 NNAY01000140 OXU30626.1 APGK01057068 KB741277 ENN71276.1 KB631617 ERL84703.1 GFDF01008770 JAV05314.1 KQ977754 KYN00069.1 KQ761768 OAD57125.1 CVRI01000067 CRL06687.1 KK856889 PTY26531.1 KQ414613 KOC68925.1 DS235418 EEB15586.1 KQ434864 KZC09031.1 CCAG010008016 QOIP01000005 RLU22353.1 KQ981864 KYN34304.1 ADTU01014740 KQ979568 KYN20831.1 KK107063 EZA61175.1 GL888818 EGI57846.1 KQ982314 KYQ57967.1 LBMM01003834 KMQ93069.1 KQ976662 KYM78489.1 GL438491 EFN68899.1 GDRN01007567 JAI67946.1 GDIQ01186460 JAK65265.1 MWRG01002896 PRD32117.1 LNIX01000028 OXA41860.1 LRGB01002011 KZS09650.1 KQ461198 KPJ05958.1 KQ459185 KPJ03356.1 GALX01007084 JAB61382.1 GDIQ01246234 JAK05491.1 KV931317 PIO31589.1 NWSH01000969 PCG73352.1 HACG01025751 CEK72616.1 GBGD01002350 JAC86539.1 KK121841 KFM81335.1 JH817213 EKC34793.1 GEZM01010125 JAV94181.1 AJFE02016426 AJFE02016427 AJFE02016428 AJFE02016429 JL621568 AEP99484.1 GDQN01003628 JAT87426.1 AACZ04054326 JH330059 EHH61502.1 AABR07045469 AABR07045470 AABR07045471 AABR07045472 KL278156 KFZ66933.1 KL419269 KFW87855.1 PPHD01006509 POI32166.1 KK483166 KFQ61127.1 KK592048 KFW00762.1 JH431704 KQ971379 EEZ97476.1 LJIG01022834 KRT78475.1 GAKP01014699 JAC44253.1 GECL01000528 JAP05596.1 AANG04003966 RQTK01000009 RUS91544.1 ADFV01164853 ADFV01164854 ADFV01164855 ADFV01164856 KK852438 KDR23898.1 AWGT02001832 OXB70419.1 NBAG03000210 PNI87629.1 AL445068 AL671518 JP007299 AER95896.1 KL232255 KFV06501.1 AQIA01049409 AHZZ02025974 ABGA01320989 ABGA01320990 ABGA01320991 ABGA01320992 ABGA01320993 NDHI03003476 PNJ39113.1 KB520915 EMP38047.1 AXCN02002066 AGTO01010121
Proteomes
UP000005204
UP000235965
UP000092462
UP000215335
UP000002358
UP000019118
+ More
UP000030742 UP000078542 UP000183832 UP000092443 UP000053825 UP000095300 UP000009046 UP000092445 UP000076502 UP000092444 UP000279307 UP000078541 UP000005205 UP000078492 UP000053097 UP000007755 UP000075809 UP000036403 UP000078540 UP000000311 UP000198287 UP000076858 UP000053240 UP000053268 UP000218220 UP000054359 UP000005408 UP000192223 UP000240080 UP000002277 UP000009130 UP000002494 UP000076420 UP000261640 UP000007266 UP000257200 UP000011712 UP000271974 UP000008225 UP000257160 UP000265080 UP000261400 UP000233140 UP000001073 UP000027135 UP000252040 UP000198419 UP000005640 UP000233120 UP000233100 UP000028761 UP000233080 UP000075901 UP000233180 UP000233200 UP000233020 UP000001595 UP000233040 UP000031443 UP000075886 UP000075881 UP000016665
UP000030742 UP000078542 UP000183832 UP000092443 UP000053825 UP000095300 UP000009046 UP000092445 UP000076502 UP000092444 UP000279307 UP000078541 UP000005205 UP000078492 UP000053097 UP000007755 UP000075809 UP000036403 UP000078540 UP000000311 UP000198287 UP000076858 UP000053240 UP000053268 UP000218220 UP000054359 UP000005408 UP000192223 UP000240080 UP000002277 UP000009130 UP000002494 UP000076420 UP000261640 UP000007266 UP000257200 UP000011712 UP000271974 UP000008225 UP000257160 UP000265080 UP000261400 UP000233140 UP000001073 UP000027135 UP000252040 UP000198419 UP000005640 UP000233120 UP000233100 UP000028761 UP000233080 UP000075901 UP000233180 UP000233200 UP000233020 UP000001595 UP000233040 UP000031443 UP000075886 UP000075881 UP000016665
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JEX6
A0A2J7R200
A0A1B0DFE9
A0A224XDZ3
A0A023F4L3
A0A232FIR0
+ More
K7J197 N6TSG8 U4TXH7 A0A1L8DG53 A0A195CJ25 A0A310SQC6 A0A1J1J2U2 A0A2R7X4G2 A0A1A9XN20 A0A0L7RDQ3 A0A1I8NU20 E0VQD0 A0A1A9ZZP9 A0A154PB20 A0A1B0FRH7 A0A3L8DQ43 A0A195F2I9 A0A158NFU1 A0A195E6W1 A0A026WYQ6 F4X6R4 A0A151XC95 A0A0J7KS73 A0A195B2R1 E2ACA9 A0A0P4WTK3 A0A0P5KBL1 A0A2P6L0K4 A0A226D9P0 A0A162EE21 A0A194QK57 A0A194QIS7 V5GIC3 A0A0P5G7C8 A0A2G9RUK5 A0A2A4JPL1 A0A0B6ZXN2 A0A069DYH3 A0A087UVE6 K1QLM6 A0A1W4X2G2 A0A1Y1N8F7 A0A2R9CDQ9 K9KCK8 A0A1E1WKN5 A0A2R9CDR7 A0A2I3TNU2 G8F2I8 A0A0G2JW64 A0A094L6E0 A0A2C9K688 A0A093QN90 A0A2P4T741 A0A091SQY4 A0A3Q3SUV7 A0A093IMA9 T1JN96 D6X3W5 A0A0T6AUB0 A0A034VRU3 A0A0V0GCK5 A0A3Q1EQA8 A0A2I2V1E3 A0A3S1AGN9 F7CR49 A0A3Q1B0G7 A0A3P8S795 A0A3B5A626 A0A2K5Z5N2 A0A2I3H9S3 A0A067RLH9 A0A341B3G8 A0A226NTM2 A0A2I3T8T6 Q5SRN1 M1EIH2 A0A093DWF8 A0A2K6BF43 A0A2K5TZC3 A0A2K5Z5S4 A0A096NFM5 A0A2K5KF60 A0A182T3I1 A0A2K6JP06 A0A2K6P181 A0A2K5C2C0 H2PK17 A0A2J8U1H4 A0A2K5PDT0 M7CBG4 A0A182QRA8 A0A2I3MLP9 A0A182K1F3 U3K871
K7J197 N6TSG8 U4TXH7 A0A1L8DG53 A0A195CJ25 A0A310SQC6 A0A1J1J2U2 A0A2R7X4G2 A0A1A9XN20 A0A0L7RDQ3 A0A1I8NU20 E0VQD0 A0A1A9ZZP9 A0A154PB20 A0A1B0FRH7 A0A3L8DQ43 A0A195F2I9 A0A158NFU1 A0A195E6W1 A0A026WYQ6 F4X6R4 A0A151XC95 A0A0J7KS73 A0A195B2R1 E2ACA9 A0A0P4WTK3 A0A0P5KBL1 A0A2P6L0K4 A0A226D9P0 A0A162EE21 A0A194QK57 A0A194QIS7 V5GIC3 A0A0P5G7C8 A0A2G9RUK5 A0A2A4JPL1 A0A0B6ZXN2 A0A069DYH3 A0A087UVE6 K1QLM6 A0A1W4X2G2 A0A1Y1N8F7 A0A2R9CDQ9 K9KCK8 A0A1E1WKN5 A0A2R9CDR7 A0A2I3TNU2 G8F2I8 A0A0G2JW64 A0A094L6E0 A0A2C9K688 A0A093QN90 A0A2P4T741 A0A091SQY4 A0A3Q3SUV7 A0A093IMA9 T1JN96 D6X3W5 A0A0T6AUB0 A0A034VRU3 A0A0V0GCK5 A0A3Q1EQA8 A0A2I2V1E3 A0A3S1AGN9 F7CR49 A0A3Q1B0G7 A0A3P8S795 A0A3B5A626 A0A2K5Z5N2 A0A2I3H9S3 A0A067RLH9 A0A341B3G8 A0A226NTM2 A0A2I3T8T6 Q5SRN1 M1EIH2 A0A093DWF8 A0A2K6BF43 A0A2K5TZC3 A0A2K5Z5S4 A0A096NFM5 A0A2K5KF60 A0A182T3I1 A0A2K6JP06 A0A2K6P181 A0A2K5C2C0 H2PK17 A0A2J8U1H4 A0A2K5PDT0 M7CBG4 A0A182QRA8 A0A2I3MLP9 A0A182K1F3 U3K871
PDB
6QDV
E-value=1.9534e-32,
Score=341
Ontologies
GO
PANTHER
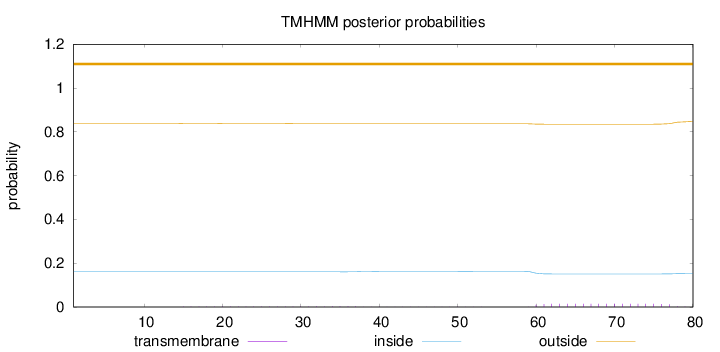
Topology
Length:
80
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30905
Exp number, first 60 AAs:
0.06983
Total prob of N-in:
0.16049
outside
1 - 80
Population Genetic Test Statistics
Pi
4.40602
Theta
9.574168
Tajima's D
-1.579898
CLR
90.63788
CSRT
0.0471476426178691
Interpretation
Uncertain
