Pre Gene Modal
BGIBMGA008072
Annotation
PREDICTED:_carbonic_anhydrase_7_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 2.036
Sequence
CDS
ATGAAGTACTTTGGTATTATAGCTGTATTCACCCTGGTGGTTATTAATGTATCTGTGGCAGCAGGATGGGGATATCGGGCCTCCGACCAGAGACGTTGGGCGGTACTGCACCCTGCTTGTGGCGGCCGTCAGCAGTCTCCCATAGCGATTTCAGCACGTCAAGCAATTCCTATCTCAATTCCAGCGATCGAACTCATTGGATACCAGAACCCTCTTCCTGGACCTTTAACGATCACTAATACGGGACATTCAGTGGCATTGACAATCCCCAAGTACACTTCGGAAGAGGAGAAGAAAGGCTTCCGTCTACCTTACATCTTTGGAGGTCCCCTAGACAACGAGTATGAGATCGATGGTCTGCATTTCCACTGGGGTGACAAGAACAACAGAGGATCGGAGCACACATTGAACGATATGCGTCTTCCTTTGGAAATGCACATCATCCATAGGAATAAGAAATACAGAAACACCGCGGAAGCTATGCAGCACCCTGATGGCCTATGTGTGCTCGCGTTCTTCTATCAGGTTGTGGAATTCGATGCCAAGCTCTTAAGTCCTATCGTGAAGAACCTAACGGCCATCGAGAACTTCAACTCCACTCTGCAGCTGCCGCATACTTTCTCCTTGTCGTCAATCCTGTCTGGCTTGGACACTGAAAGGTTCTACACGTACAAAGGATCATTGACCACTCCTCCCTGTGCTGAAGCCGTCACATGGGTGATCTTTTCTGACTACCTGCCCATTTCCGTGTTTCAAATGGATAATTTCCGTGGTCTTCTTTCCAATCTCAATTTGCCATTAGTTGATAACTTCAGGCAGTTACAACCGTTGTTTGGTCGCCGTGTCTTCGTCAGAATCACATCAAAGAACCCTAAGTTCAAGAAGACCAAACTCCACTACTCCAAATGGGACTGGGTTGGACACAAGAAGTCCGAAAATGACGTCATCGACTTCGATGAATAA
Protein
MKYFGIIAVFTLVVINVSVAAGWGYRASDQRRWAVLHPACGGRQQSPIAISARQAIPISIPAIELIGYQNPLPGPLTITNTGHSVALTIPKYTSEEEKKGFRLPYIFGGPLDNEYEIDGLHFHWGDKNNRGSEHTLNDMRLPLEMHIIHRNKKYRNTAEAMQHPDGLCVLAFFYQVVEFDAKLLSPIVKNLTAIENFNSTLQLPHTFSLSSILSGLDTERFYTYKGSLTTPPCAEAVTWVIFSDYLPISVFQMDNFRGLLSNLNLPLVDNFRQLQPLFGRRVFVRITSKNPKFKKTKLHYSKWDWVGHKKSENDVIDFDE
Summary
Similarity
Belongs to the alpha-carbonic anhydrase family.
Uniprot
H9JEX5
A0A3S2LTP0
A0A2A4JMQ5
A0A2W1BGF7
A0A212F6P0
A0A194QCU0
+ More
A0A2H1W7W3 A0A194QLX2 A0A0L7K418 A0A0L7LNP0 A0A2A4JP43 A0A2M4A4D7 A0A2M4A4B8 A0A1B0D305 A0A2M4BU27 W5J5H4 A0A2M4BUJ3 A0A2M4BUI5 A0A2M3Z0B7 A0A084VNM6 A0A2M3Z0B9 A0A182QWW4 A0A182FQQ2 A0A182RHX5 A0A182MCM5 A0A182NL58 A0A182VQM4 A0A182P5S0 Q7QKJ5 A0A023EPC9 A0A182HJF2 U5ESR7 A0A182JQ77 A0A182X1Y4 A0A182VGI8 Q16XM2 A0A182IS97 A0A023EN84 A0A182LCR1 Q19MS2 A0A182THY7 A0A182YK91 A0A182HEM3 A0A2M3ZET7 A0A0K8WDE1 A0A034W935 W8ADY5 A0A0K8UFR7 A0A034WCQ3 A0A0L0BMT7 A0A0M4EH41 Q29AD2 A0A1I8MB82 A0A1A9YDP1 W8APD1 T1PHX8 B3MT23 B4QXE4 B4IC41 A0A1I8Q6R1 A0A1L8EG69 B4K9Q3 Q9VB76 B4M109 H1UU99 B3P858 B4JHF6 A0A1B0G191 B4PRV3 Q6NLI8 A0A1W4U5W2 B4N8D8 A0A1A9UTG9 A0A1A9W9J3 A0A1J1HET1 A0A1B0B318 A0A2M3Z2B0 B4G2F9 A0A1A9ZSR2 A0A0K8UTW3 A0A1B6CUQ3 A0A1B6J557 A0A1B6LJH5 A0A1B6GV12 A0A1W4WVX4 A0A087ZQS3 A0A0C9R1G8 A0A0N0U3E9 A0A224XTH0 A0A2A3EJI8 A0A3B0JDX5 A0A0T6AWA5 A0A3L8DSQ2 A0A0A9YFG2 A0A023F0Q2 A0A154PUP8 A0A310S8K5 A0A2P8XJ35 A0A2S1TNS1
A0A2H1W7W3 A0A194QLX2 A0A0L7K418 A0A0L7LNP0 A0A2A4JP43 A0A2M4A4D7 A0A2M4A4B8 A0A1B0D305 A0A2M4BU27 W5J5H4 A0A2M4BUJ3 A0A2M4BUI5 A0A2M3Z0B7 A0A084VNM6 A0A2M3Z0B9 A0A182QWW4 A0A182FQQ2 A0A182RHX5 A0A182MCM5 A0A182NL58 A0A182VQM4 A0A182P5S0 Q7QKJ5 A0A023EPC9 A0A182HJF2 U5ESR7 A0A182JQ77 A0A182X1Y4 A0A182VGI8 Q16XM2 A0A182IS97 A0A023EN84 A0A182LCR1 Q19MS2 A0A182THY7 A0A182YK91 A0A182HEM3 A0A2M3ZET7 A0A0K8WDE1 A0A034W935 W8ADY5 A0A0K8UFR7 A0A034WCQ3 A0A0L0BMT7 A0A0M4EH41 Q29AD2 A0A1I8MB82 A0A1A9YDP1 W8APD1 T1PHX8 B3MT23 B4QXE4 B4IC41 A0A1I8Q6R1 A0A1L8EG69 B4K9Q3 Q9VB76 B4M109 H1UU99 B3P858 B4JHF6 A0A1B0G191 B4PRV3 Q6NLI8 A0A1W4U5W2 B4N8D8 A0A1A9UTG9 A0A1A9W9J3 A0A1J1HET1 A0A1B0B318 A0A2M3Z2B0 B4G2F9 A0A1A9ZSR2 A0A0K8UTW3 A0A1B6CUQ3 A0A1B6J557 A0A1B6LJH5 A0A1B6GV12 A0A1W4WVX4 A0A087ZQS3 A0A0C9R1G8 A0A0N0U3E9 A0A224XTH0 A0A2A3EJI8 A0A3B0JDX5 A0A0T6AWA5 A0A3L8DSQ2 A0A0A9YFG2 A0A023F0Q2 A0A154PUP8 A0A310S8K5 A0A2P8XJ35 A0A2S1TNS1
Pubmed
19121390
28756777
22118469
26354079
26227816
20920257
+ More
23761445 24438588 12364791 14747013 17210077 24945155 17510324 20966253 25244985 26483478 25348373 24495485 26108605 15632085 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 30249741 25401762 26823975 25474469 29403074 29727440
23761445 24438588 12364791 14747013 17210077 24945155 17510324 20966253 25244985 26483478 25348373 24495485 26108605 15632085 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 30249741 25401762 26823975 25474469 29403074 29727440
EMBL
BABH01023425
BABH01023426
BABH01023427
RSAL01000211
RVE44197.1
NWSH01000969
+ More
PCG73355.1 KZ150199 PZC72287.1 AGBW02009996 OWR49379.1 KQ459185 KPJ03358.1 ODYU01006897 SOQ49160.1 KQ461198 KPJ05960.1 JTDY01011745 KOB53514.1 JTDY01000482 KOB76979.1 PCG73354.1 GGFK01002290 MBW35611.1 GGFK01002270 MBW35591.1 AJVK01010815 GGFJ01007434 MBW56575.1 ADMH02002181 ETN58014.1 GGFJ01007532 MBW56673.1 GGFJ01007533 MBW56674.1 GGFM01001212 MBW21963.1 ATLV01014774 KE524988 KFB39570.1 GGFM01001214 MBW21965.1 AXCN02000083 AXCM01001920 AAAB01008794 EAA03529.3 GAPW01002748 GAPW01002747 GAPW01002745 JAC10851.1 APCN01002159 GANO01003132 JAB56739.1 CH477536 EAT39369.1 GAPW01002746 JAC10852.1 DQ518577 ABF66619.1 JXUM01035638 JXUM01035639 KQ561068 KXJ79806.1 GGFM01006336 MBW27087.1 GDHF01003464 JAI48850.1 GAKP01006856 JAC52096.1 GAMC01020186 GAMC01020185 JAB86370.1 GDHF01026797 JAI25517.1 GAKP01006855 JAC52097.1 JRES01001712 KNC20579.1 CP012526 ALC45584.1 CM000070 EAL27418.1 GAMC01020189 GAMC01020187 JAB86368.1 KA647735 AFP62364.1 CH902623 EDV30413.1 CM000364 EDX14630.1 CH480828 EDW45199.1 GFDG01001072 JAV17727.1 CH933806 EDW14528.1 AE014297 AAF56666.2 CH940650 EDW67420.1 BT132983 AEX31653.1 CH954182 EDV53462.1 CH916369 EDV92783.1 CCAG010001934 CM000160 EDW98546.1 BT012344 AAS77469.1 CH964232 EDW81389.1 CVRI01000001 CRK86501.1 JXJN01007759 GGFM01001890 MBW22641.1 CH479179 EDW24004.1 GDHF01022175 JAI30139.1 GEDC01020082 GEDC01006389 GEDC01001615 JAS17216.1 JAS30909.1 JAS35683.1 GECU01013409 JAS94297.1 GEBQ01016218 JAT23759.1 GECZ01003489 JAS66280.1 GBYB01000706 JAG70473.1 KQ435911 KOX68932.1 GFTR01004604 JAW11822.1 KZ288226 PBC31868.1 OUUW01000005 SPP80564.1 LJIG01022703 KRT79149.1 QOIP01000004 RLU23356.1 GBHO01018343 GBHO01018342 GBHO01018341 GBHO01013756 GBHO01013755 GBHO01008619 GBHO01008618 GBHO01008616 GBHO01008615 GBHO01008611 GBRD01012103 GBRD01010966 GBRD01010964 GBRD01010963 GDHC01018896 GDHC01014892 GDHC01013354 JAG25261.1 JAG25262.1 JAG25263.1 JAG29848.1 JAG29849.1 JAG34985.1 JAG34986.1 JAG34988.1 JAG34989.1 JAG34993.1 JAG53721.1 JAP99732.1 JAQ03737.1 JAQ05275.1 GBBI01004136 JAC14576.1 KQ435207 KZC15078.1 KQ769179 OAD52958.1 PYGN01001954 PSN32015.1 MF189029 AWI63383.1
PCG73355.1 KZ150199 PZC72287.1 AGBW02009996 OWR49379.1 KQ459185 KPJ03358.1 ODYU01006897 SOQ49160.1 KQ461198 KPJ05960.1 JTDY01011745 KOB53514.1 JTDY01000482 KOB76979.1 PCG73354.1 GGFK01002290 MBW35611.1 GGFK01002270 MBW35591.1 AJVK01010815 GGFJ01007434 MBW56575.1 ADMH02002181 ETN58014.1 GGFJ01007532 MBW56673.1 GGFJ01007533 MBW56674.1 GGFM01001212 MBW21963.1 ATLV01014774 KE524988 KFB39570.1 GGFM01001214 MBW21965.1 AXCN02000083 AXCM01001920 AAAB01008794 EAA03529.3 GAPW01002748 GAPW01002747 GAPW01002745 JAC10851.1 APCN01002159 GANO01003132 JAB56739.1 CH477536 EAT39369.1 GAPW01002746 JAC10852.1 DQ518577 ABF66619.1 JXUM01035638 JXUM01035639 KQ561068 KXJ79806.1 GGFM01006336 MBW27087.1 GDHF01003464 JAI48850.1 GAKP01006856 JAC52096.1 GAMC01020186 GAMC01020185 JAB86370.1 GDHF01026797 JAI25517.1 GAKP01006855 JAC52097.1 JRES01001712 KNC20579.1 CP012526 ALC45584.1 CM000070 EAL27418.1 GAMC01020189 GAMC01020187 JAB86368.1 KA647735 AFP62364.1 CH902623 EDV30413.1 CM000364 EDX14630.1 CH480828 EDW45199.1 GFDG01001072 JAV17727.1 CH933806 EDW14528.1 AE014297 AAF56666.2 CH940650 EDW67420.1 BT132983 AEX31653.1 CH954182 EDV53462.1 CH916369 EDV92783.1 CCAG010001934 CM000160 EDW98546.1 BT012344 AAS77469.1 CH964232 EDW81389.1 CVRI01000001 CRK86501.1 JXJN01007759 GGFM01001890 MBW22641.1 CH479179 EDW24004.1 GDHF01022175 JAI30139.1 GEDC01020082 GEDC01006389 GEDC01001615 JAS17216.1 JAS30909.1 JAS35683.1 GECU01013409 JAS94297.1 GEBQ01016218 JAT23759.1 GECZ01003489 JAS66280.1 GBYB01000706 JAG70473.1 KQ435911 KOX68932.1 GFTR01004604 JAW11822.1 KZ288226 PBC31868.1 OUUW01000005 SPP80564.1 LJIG01022703 KRT79149.1 QOIP01000004 RLU23356.1 GBHO01018343 GBHO01018342 GBHO01018341 GBHO01013756 GBHO01013755 GBHO01008619 GBHO01008618 GBHO01008616 GBHO01008615 GBHO01008611 GBRD01012103 GBRD01010966 GBRD01010964 GBRD01010963 GDHC01018896 GDHC01014892 GDHC01013354 JAG25261.1 JAG25262.1 JAG25263.1 JAG29848.1 JAG29849.1 JAG34985.1 JAG34986.1 JAG34988.1 JAG34989.1 JAG34993.1 JAG53721.1 JAP99732.1 JAQ03737.1 JAQ05275.1 GBBI01004136 JAC14576.1 KQ435207 KZC15078.1 KQ769179 OAD52958.1 PYGN01001954 PSN32015.1 MF189029 AWI63383.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000037510 UP000092462 UP000000673 UP000030765 UP000075886 UP000069272 UP000075900 UP000075883 UP000075884 UP000075920 UP000075885 UP000007062 UP000075840 UP000075881 UP000076407 UP000075903 UP000008820 UP000075880 UP000075882 UP000075902 UP000076408 UP000069940 UP000249989 UP000037069 UP000092553 UP000001819 UP000095301 UP000092443 UP000007801 UP000000304 UP000001292 UP000095300 UP000009192 UP000000803 UP000008792 UP000008711 UP000001070 UP000092444 UP000002282 UP000192221 UP000007798 UP000078200 UP000091820 UP000183832 UP000092460 UP000008744 UP000092445 UP000192223 UP000005203 UP000053105 UP000242457 UP000268350 UP000279307 UP000076502 UP000245037
UP000037510 UP000092462 UP000000673 UP000030765 UP000075886 UP000069272 UP000075900 UP000075883 UP000075884 UP000075920 UP000075885 UP000007062 UP000075840 UP000075881 UP000076407 UP000075903 UP000008820 UP000075880 UP000075882 UP000075902 UP000076408 UP000069940 UP000249989 UP000037069 UP000092553 UP000001819 UP000095301 UP000092443 UP000007801 UP000000304 UP000001292 UP000095300 UP000009192 UP000000803 UP000008792 UP000008711 UP000001070 UP000092444 UP000002282 UP000192221 UP000007798 UP000078200 UP000091820 UP000183832 UP000092460 UP000008744 UP000092445 UP000192223 UP000005203 UP000053105 UP000242457 UP000268350 UP000279307 UP000076502 UP000245037
Interpro
Gene 3D
ProteinModelPortal
H9JEX5
A0A3S2LTP0
A0A2A4JMQ5
A0A2W1BGF7
A0A212F6P0
A0A194QCU0
+ More
A0A2H1W7W3 A0A194QLX2 A0A0L7K418 A0A0L7LNP0 A0A2A4JP43 A0A2M4A4D7 A0A2M4A4B8 A0A1B0D305 A0A2M4BU27 W5J5H4 A0A2M4BUJ3 A0A2M4BUI5 A0A2M3Z0B7 A0A084VNM6 A0A2M3Z0B9 A0A182QWW4 A0A182FQQ2 A0A182RHX5 A0A182MCM5 A0A182NL58 A0A182VQM4 A0A182P5S0 Q7QKJ5 A0A023EPC9 A0A182HJF2 U5ESR7 A0A182JQ77 A0A182X1Y4 A0A182VGI8 Q16XM2 A0A182IS97 A0A023EN84 A0A182LCR1 Q19MS2 A0A182THY7 A0A182YK91 A0A182HEM3 A0A2M3ZET7 A0A0K8WDE1 A0A034W935 W8ADY5 A0A0K8UFR7 A0A034WCQ3 A0A0L0BMT7 A0A0M4EH41 Q29AD2 A0A1I8MB82 A0A1A9YDP1 W8APD1 T1PHX8 B3MT23 B4QXE4 B4IC41 A0A1I8Q6R1 A0A1L8EG69 B4K9Q3 Q9VB76 B4M109 H1UU99 B3P858 B4JHF6 A0A1B0G191 B4PRV3 Q6NLI8 A0A1W4U5W2 B4N8D8 A0A1A9UTG9 A0A1A9W9J3 A0A1J1HET1 A0A1B0B318 A0A2M3Z2B0 B4G2F9 A0A1A9ZSR2 A0A0K8UTW3 A0A1B6CUQ3 A0A1B6J557 A0A1B6LJH5 A0A1B6GV12 A0A1W4WVX4 A0A087ZQS3 A0A0C9R1G8 A0A0N0U3E9 A0A224XTH0 A0A2A3EJI8 A0A3B0JDX5 A0A0T6AWA5 A0A3L8DSQ2 A0A0A9YFG2 A0A023F0Q2 A0A154PUP8 A0A310S8K5 A0A2P8XJ35 A0A2S1TNS1
A0A2H1W7W3 A0A194QLX2 A0A0L7K418 A0A0L7LNP0 A0A2A4JP43 A0A2M4A4D7 A0A2M4A4B8 A0A1B0D305 A0A2M4BU27 W5J5H4 A0A2M4BUJ3 A0A2M4BUI5 A0A2M3Z0B7 A0A084VNM6 A0A2M3Z0B9 A0A182QWW4 A0A182FQQ2 A0A182RHX5 A0A182MCM5 A0A182NL58 A0A182VQM4 A0A182P5S0 Q7QKJ5 A0A023EPC9 A0A182HJF2 U5ESR7 A0A182JQ77 A0A182X1Y4 A0A182VGI8 Q16XM2 A0A182IS97 A0A023EN84 A0A182LCR1 Q19MS2 A0A182THY7 A0A182YK91 A0A182HEM3 A0A2M3ZET7 A0A0K8WDE1 A0A034W935 W8ADY5 A0A0K8UFR7 A0A034WCQ3 A0A0L0BMT7 A0A0M4EH41 Q29AD2 A0A1I8MB82 A0A1A9YDP1 W8APD1 T1PHX8 B3MT23 B4QXE4 B4IC41 A0A1I8Q6R1 A0A1L8EG69 B4K9Q3 Q9VB76 B4M109 H1UU99 B3P858 B4JHF6 A0A1B0G191 B4PRV3 Q6NLI8 A0A1W4U5W2 B4N8D8 A0A1A9UTG9 A0A1A9W9J3 A0A1J1HET1 A0A1B0B318 A0A2M3Z2B0 B4G2F9 A0A1A9ZSR2 A0A0K8UTW3 A0A1B6CUQ3 A0A1B6J557 A0A1B6LJH5 A0A1B6GV12 A0A1W4WVX4 A0A087ZQS3 A0A0C9R1G8 A0A0N0U3E9 A0A224XTH0 A0A2A3EJI8 A0A3B0JDX5 A0A0T6AWA5 A0A3L8DSQ2 A0A0A9YFG2 A0A023F0Q2 A0A154PUP8 A0A310S8K5 A0A2P8XJ35 A0A2S1TNS1
PDB
6H38
E-value=6.33314e-35,
Score=368
Ontologies
PATHWAY
PANTHER
Topology
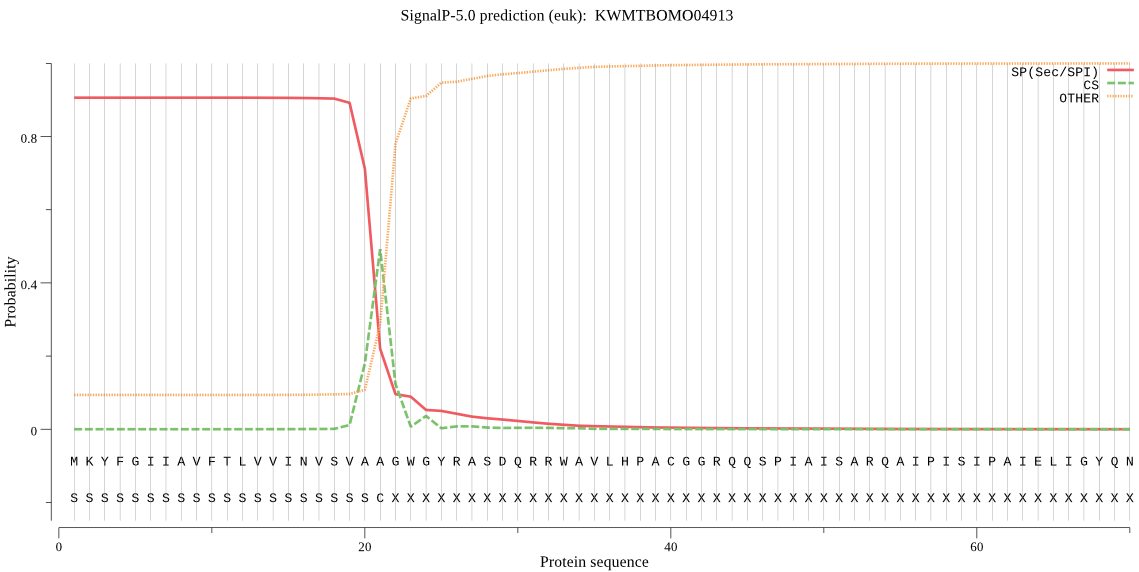
SignalP
Position: 1 - 21,
Likelihood: 0.905646
Length:
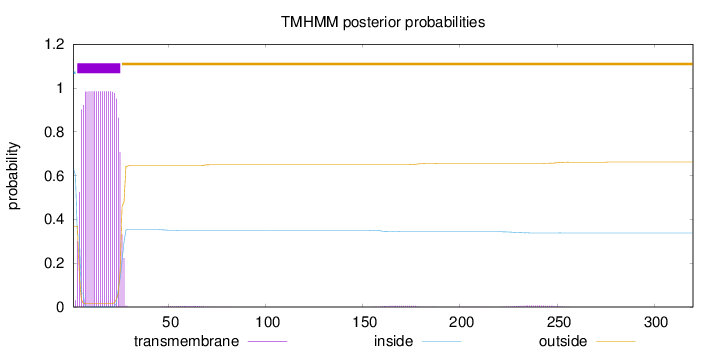
320
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.85102
Exp number, first 60 AAs:
21.55849
Total prob of N-in:
0.63103
POSSIBLE N-term signal
sequence
inside
1 - 2
TMhelix
3 - 25
outside
26 - 320
Population Genetic Test Statistics
Pi
187.383642
Theta
17.648133
Tajima's D
-0.841232
CLR
1.312311
CSRT
0.165641717914104
Interpretation
Uncertain
