Pre Gene Modal
BGIBMGA008013
Annotation
Protein_fat-free-like_protein_[Operophtera_brumata]
Full name
Vacuolar protein sorting-associated protein 51 homolog
Alternative Name
Protein fat-free homolog
Location in the cell
Nuclear Reliability : 2.04
Sequence
CDS
ATGGAGGAAGCCAAAGCGGTTAAAGAAAATCCTCTGGACATCGACAGCAGTAGTTTCAGCCCAGACGTCTACTTAGATTGTCTTCTAAAATGCGCTACCCTAAAGCAAGTAATGGACAAAGAAGCGGAAATTGTAACTCAAAGTCAATTTCTACAATCAGAAATGCAGACATTGGTTTATGAAAATTATAATAAATTTATTTCGGCGACCGAAACTGTACGCAAAATGCGTTCCGATTTTAAAATAATGCAAGAAGAAATGAATAAACTTAGCGAGAATATCAATAAAATAACTACATTCAGCTCTCAAATATCTGAAAATCTCCAAGGCAGCGGTAATAATGTTCGACGTTTGTGTGGTACAAGACAGCTGTTAGATAAACTTCAGTTCCTTTTCCTTCTACCTTCTCAACTTAACAAAGCAATAGCAGAAGGCAGATACAGTGATGCTGTACAAGACTATTCACATGCTCAGCGAGTACTCCAAAAATATGGCAATCAGCCTTCATTCCAAAGCATACAGACTGAGTGTTCAGAAATTATATTTGAACTGAAGAAGACACTCCGTGAAAGACTTATCAGTCCTAATACATCTGCAACTGAACTTGCTGAAAGTGTAGCCCTTTTAAGGCAGCTCCAAGAAAGTGATTCATCCTTACAAGATATATTCCTTAGTTGTGCAGAGAGTCGTCTTGACAAACACTTGCAGATGCTCAGTTCTATAGCTGAACATACAGACATTTTGGACTGGGTAGAAAAATGCAATAATACATTGTTGGCTGATTTAGCTATTATAATATCATGTTACAATGAAATGTTCGAACAGGAAGCTATCATACCCGAATTTGCTGATAAAATAATGATTCAAATTTTTCATTTATTCGAAGAGGTCATTAAAAGACCTGAGAACAACGGAACTGAAATGTTAATAAGGGGCCTTGATAAATTCTTCAGAAAACTCCAGGCTATGACCGAAATATCATCAAGTGATATGATGTCCAGTAAAGGCACTGAGGTTGTCATTCAGTGTATAAATCACAAGGCTATGGTACAGTACCAGAGCATACAGACTCAATTTAAAGACAGTCTTATGAAAGTAAGGCAGTCTTTGGCCAGTAAAGGATCAGAAACAATTGACTTAAAAGATATACTGAACTCCCTTCAAGTATATCTGATGCAAAAAATCCAAGCCTCACTCTTAGATCTAATGGCATTCTTACAAAATAATTTATCATTTGGATTAAAACCTTGGTGTGCAAAAGCAGTAGCTGATGCTTGCTCAAAAGTCATAAATGAAACTATGGCTAATCTTTCTGATCTGGTAAAAAAGATGGCTTGCCTTCAGACTTCAAATAACAACATTCCTTTTGAATTGCTACTTATACTAGCTAAGCTATGTTTGGATATGCAAGAGACTGGTGTCTCGACATTACAGAACCATCTTCTCAAACTCTTAGAAGAGGCTGCTCCTGGCCTCACTGTGGATAACAAGGATACTAGTAGTGTCATGGTCCACTTATCTACTGCAGCGCAGACTGCACTAGATTCAGAAGTTGTATTTATTGGCCAGACTGCAGCCCAAATGTTGAGAGTATCTGTATTAGCTAGAGATTGGTTGCGAGCTCCAGAACCAAGGGGGCCAAGAGCAGTATGCAGAAGAGTAGTGGAGACCTTAGCTAGTGCTGATGGTGCTGCATCTCAACTGTTTCCAACTAGTATTAAACCTTCTAGTGACTCCAGTCGGCGTACTATTTGGTCACGTGCTCCTTCTTCTTTTTCACCTCTTAATAGGATATTCTCTGAAAGAATTGAAGTGTTTAGTCCTGCAGGGGCAGACAGAGCTGCACTATCAAATGGTGCCCTAAAAGTAGCTTTGAAAGCATTAGTTGAATGTGTGCGCCTACGCACATTTGGAAGGCATGGCCTACAGCAATTACAAGTTGATGTTCACTTTCTACAGCAAAGGCTTTCATGTATGGGCAATGATGAGAGGCTTCTAAATGCTTTGCTTGAAGATGCTTTAGCATCAGCACAGTTGAGATGCGTTGACCCACAACTAATGGAACCTAGTATTGTAGATATTATTTGTGAAAGAGGTTGA
Protein
MEEAKAVKENPLDIDSSSFSPDVYLDCLLKCATLKQVMDKEAEIVTQSQFLQSEMQTLVYENYNKFISATETVRKMRSDFKIMQEEMNKLSENINKITTFSSQISENLQGSGNNVRRLCGTRQLLDKLQFLFLLPSQLNKAIAEGRYSDAVQDYSHAQRVLQKYGNQPSFQSIQTECSEIIFELKKTLRERLISPNTSATELAESVALLRQLQESDSSLQDIFLSCAESRLDKHLQMLSSIAEHTDILDWVEKCNNTLLADLAIIISCYNEMFEQEAIIPEFADKIMIQIFHLFEEVIKRPENNGTEMLIRGLDKFFRKLQAMTEISSSDMMSSKGTEVVIQCINHKAMVQYQSIQTQFKDSLMKVRQSLASKGSETIDLKDILNSLQVYLMQKIQASLLDLMAFLQNNLSFGLKPWCAKAVADACSKVINETMANLSDLVKKMACLQTSNNNIPFELLLILAKLCLDMQETGVSTLQNHLLKLLEEAAPGLTVDNKDTSSVMVHLSTAAQTALDSEVVFIGQTAAQMLRVSVLARDWLRAPEPRGPRAVCRRVVETLASADGAASQLFPTSIKPSSDSSRRTIWSRAPSSFSPLNRIFSERIEVFSPAGADRAALSNGALKVALKALVECVRLRTFGRHGLQQLQVDVHFLQQRLSCMGNDERLLNALLEDALASAQLRCVDPQLMEPSIVDIICERG
Summary
Description
May act as a component of the GARP complex that is involved in retrograde transport from early and late endosomes to the trans-Golgi network (TGN).
Involved in retrograde transport from early and late endosomes to the late Golgi. The GARP complex is required for the maintenance of protein retrieval from endosomes to the TGN, acid hydrolase sorting, lysosome function, endosomal cholesterol traffic and autophagy. Acts as component of the EARP complex that is involved in endocytic recycling.
Involved in retrograde transport from early and late endosomes to the late Golgi. The GARP complex is required for the maintenance of protein retrieval from endosomes to the TGN, acid hydrolase sorting, lysosome function, endosomal cholesterol traffic and autophagy. Acts as component of the EARP complex that is involved in endocytic recycling.
Subunit
Component of the Golgi-associated retrograde protein (GARP) complex.
Component of the Golgi-associated retrograde protein (GARP) complex Component of the endosome-associated retrograde protein (EARP) complex.
Component of the Golgi-associated retrograde protein (GARP) complex Component of the endosome-associated retrograde protein (EARP) complex.
Similarity
Belongs to the VPS51 family.
Keywords
Coiled coil
Complete proteome
Reference proteome
Endosome
Golgi apparatus
Lipid transport
Protein transport
Transport
Feature
chain Vacuolar protein sorting-associated protein 51 homolog
Uniprot
H9JER6
A0A0L7LNG8
A0A2H1WXU1
A0A2W1BD04
A0A2A4JNG2
A0A194QK63
+ More
A0A194QIT2 A0A1E1W774 B0X770 A0A1Q3F808 A0A0N0BEZ7 A0A067QJH8 A0A088AF26 A0A2A3EV14 A0A2J7RFQ4 A0A232FNE9 A0A195BBN4 K7INY4 A0A151JCS4 A0A195FLH2 Q16KJ4 A0A182HB46 A0A1Y1ND64 A0A151IQH1 E2B9H0 A0A0C9PUE1 A0A3L8DDX5 A0A336LXL0 Q7Q6Z6 B4LJ46 D6X272 A0A182U4T4 A0A1S4FXK0 A0A1S3IN33 A0A182MUN4 A0A182RMJ1 A0A182XN22 B4J7M9 A0A182WKG8 A0A182LJA6 A0A0L7QYD5 A0A1B6MLS5 A0A182VH94 B3MI33 A0A1B6GXN5 A0A0M4EKU1 A0A182HWX9 B4KPL2 A0A1W4UAZ0 A0A1A9UY46 A0A2C9K343 A0A3B0J2W4 B4QD20 Q8MSY4 W8BZL6 B4HP05 A0A1W4UP77 A0A0L0CHZ7 B3NKA2 B4P4A1 A0A1A9Y0G8 Q291X2 B4GA52 A0A1B0B676 A0A0A1XN24 A0A1B0CB26 A0A0K8U7X5 A0A182FEH8 E0VIJ9 W5J5F2 B4MK00 A0A182NHQ4 A0A0K8U6Z8 A0A1J1J2L3 A0A1I8N657 A0A1I8N671 N6TX68 A0A2J7RFP7 A0A026WSA8 A0A1I8N667 A0A2S2NRF4 J9K7Q5 V4CEQ7 A0A0B6ZII1 A0A2H8TG56 A0A1A9ZY13 A0A1I8QD32 Q505L3 A0A1L8GCZ5 R7V5S5 F4WR83 A0A1I8QD70 Q4V9Y0 A0A1L8GJL8 A0A3B0IZ17 A0A3P9LE30 T1J0Z4 A0A1E1XI44
A0A194QIT2 A0A1E1W774 B0X770 A0A1Q3F808 A0A0N0BEZ7 A0A067QJH8 A0A088AF26 A0A2A3EV14 A0A2J7RFQ4 A0A232FNE9 A0A195BBN4 K7INY4 A0A151JCS4 A0A195FLH2 Q16KJ4 A0A182HB46 A0A1Y1ND64 A0A151IQH1 E2B9H0 A0A0C9PUE1 A0A3L8DDX5 A0A336LXL0 Q7Q6Z6 B4LJ46 D6X272 A0A182U4T4 A0A1S4FXK0 A0A1S3IN33 A0A182MUN4 A0A182RMJ1 A0A182XN22 B4J7M9 A0A182WKG8 A0A182LJA6 A0A0L7QYD5 A0A1B6MLS5 A0A182VH94 B3MI33 A0A1B6GXN5 A0A0M4EKU1 A0A182HWX9 B4KPL2 A0A1W4UAZ0 A0A1A9UY46 A0A2C9K343 A0A3B0J2W4 B4QD20 Q8MSY4 W8BZL6 B4HP05 A0A1W4UP77 A0A0L0CHZ7 B3NKA2 B4P4A1 A0A1A9Y0G8 Q291X2 B4GA52 A0A1B0B676 A0A0A1XN24 A0A1B0CB26 A0A0K8U7X5 A0A182FEH8 E0VIJ9 W5J5F2 B4MK00 A0A182NHQ4 A0A0K8U6Z8 A0A1J1J2L3 A0A1I8N657 A0A1I8N671 N6TX68 A0A2J7RFP7 A0A026WSA8 A0A1I8N667 A0A2S2NRF4 J9K7Q5 V4CEQ7 A0A0B6ZII1 A0A2H8TG56 A0A1A9ZY13 A0A1I8QD32 Q505L3 A0A1L8GCZ5 R7V5S5 F4WR83 A0A1I8QD70 Q4V9Y0 A0A1L8GJL8 A0A3B0IZ17 A0A3P9LE30 T1J0Z4 A0A1E1XI44
Pubmed
19121390
26227816
28756777
26354079
24845553
28648823
+ More
20075255 17510324 26483478 28004739 20798317 30249741 12364791 17994087 18362917 19820115 20966253 15562597 22936249 10731132 12537572 12537569 24495485 26108605 17550304 15632085 25830018 20566863 20920257 23761445 25315136 23537049 24508170 23254933 27762356 21719571 17554307 28503490
20075255 17510324 26483478 28004739 20798317 30249741 12364791 17994087 18362917 19820115 20966253 15562597 22936249 10731132 12537572 12537569 24495485 26108605 17550304 15632085 25830018 20566863 20920257 23761445 25315136 23537049 24508170 23254933 27762356 21719571 17554307 28503490
EMBL
BABH01023425
JTDY01000482
KOB76980.1
ODYU01011873
SOQ57868.1
KZ150199
+ More
PZC72291.1 NWSH01000969 PCG73359.1 KQ461198 KPJ05963.1 KQ459185 KPJ03361.1 GDQN01008353 JAT82701.1 DS232440 EDS41764.1 GFDL01011340 JAV23705.1 KQ435821 KOX72348.1 KK853273 KDR09124.1 KZ288185 PBC34911.1 NEVH01004410 PNF39658.1 NNAY01000018 OXU31897.1 KQ976532 KYM81620.1 KQ979048 KYN23217.1 KQ981512 KYN40839.1 CH477954 EAT34821.1 JXUM01031693 KQ560932 KXJ80293.1 GEZM01006009 JAV95833.1 KQ976780 KYN08398.1 GL446527 EFN87653.1 GBYB01004988 JAG74755.1 QOIP01000009 RLU18531.1 UFQS01000205 UFQT01000205 SSX01331.1 SSX21711.1 AAAB01008960 EAA11747.3 CH940648 EDW61482.2 KQ971371 EFA10222.1 AXCM01016594 CH916367 EDW02177.1 KQ414688 KOC63630.1 GEBQ01003125 JAT36852.1 CH902619 EDV38043.1 GECZ01002590 JAS67179.1 CP012524 ALC42067.1 APCN01001587 CH933808 EDW09122.1 OUUW01000001 SPP73503.1 CM000362 CM002911 EDX07725.1 KMY94924.1 AE013599 AY118494 GAMC01007794 JAB98761.1 CH480816 EDW48507.1 JRES01000378 KNC31837.1 CH954179 EDV55124.1 CM000158 EDW91587.1 CM000071 EAL24990.1 CH479181 EDW31804.1 JXJN01009007 GBXI01002022 JAD12270.1 AJWK01004777 GDHF01029633 JAI22681.1 DS235200 EEB13205.1 ADMH02002097 ETN59216.1 CH963846 EDW72439.1 GDHF01029961 JAI22353.1 CVRI01000064 CRL05073.1 APGK01057453 KB741280 KB632033 ENN70912.1 ERL88212.1 PNF39659.1 KK107119 EZA58546.1 GGMR01007151 MBY19770.1 ABLF02022130 KB200702 ESP00450.1 HACG01021544 CEK68409.1 GFXV01001259 MBW13064.1 BC094495 CM004473 OCT81748.1 AMQN01004913 KB294684 ELU14198.1 GL888284 EGI63300.1 BC096636 CM004472 OCT84019.1 SPP73504.1 JH431742 GFAC01000432 JAT98756.1
PZC72291.1 NWSH01000969 PCG73359.1 KQ461198 KPJ05963.1 KQ459185 KPJ03361.1 GDQN01008353 JAT82701.1 DS232440 EDS41764.1 GFDL01011340 JAV23705.1 KQ435821 KOX72348.1 KK853273 KDR09124.1 KZ288185 PBC34911.1 NEVH01004410 PNF39658.1 NNAY01000018 OXU31897.1 KQ976532 KYM81620.1 KQ979048 KYN23217.1 KQ981512 KYN40839.1 CH477954 EAT34821.1 JXUM01031693 KQ560932 KXJ80293.1 GEZM01006009 JAV95833.1 KQ976780 KYN08398.1 GL446527 EFN87653.1 GBYB01004988 JAG74755.1 QOIP01000009 RLU18531.1 UFQS01000205 UFQT01000205 SSX01331.1 SSX21711.1 AAAB01008960 EAA11747.3 CH940648 EDW61482.2 KQ971371 EFA10222.1 AXCM01016594 CH916367 EDW02177.1 KQ414688 KOC63630.1 GEBQ01003125 JAT36852.1 CH902619 EDV38043.1 GECZ01002590 JAS67179.1 CP012524 ALC42067.1 APCN01001587 CH933808 EDW09122.1 OUUW01000001 SPP73503.1 CM000362 CM002911 EDX07725.1 KMY94924.1 AE013599 AY118494 GAMC01007794 JAB98761.1 CH480816 EDW48507.1 JRES01000378 KNC31837.1 CH954179 EDV55124.1 CM000158 EDW91587.1 CM000071 EAL24990.1 CH479181 EDW31804.1 JXJN01009007 GBXI01002022 JAD12270.1 AJWK01004777 GDHF01029633 JAI22681.1 DS235200 EEB13205.1 ADMH02002097 ETN59216.1 CH963846 EDW72439.1 GDHF01029961 JAI22353.1 CVRI01000064 CRL05073.1 APGK01057453 KB741280 KB632033 ENN70912.1 ERL88212.1 PNF39659.1 KK107119 EZA58546.1 GGMR01007151 MBY19770.1 ABLF02022130 KB200702 ESP00450.1 HACG01021544 CEK68409.1 GFXV01001259 MBW13064.1 BC094495 CM004473 OCT81748.1 AMQN01004913 KB294684 ELU14198.1 GL888284 EGI63300.1 BC096636 CM004472 OCT84019.1 SPP73504.1 JH431742 GFAC01000432 JAT98756.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000002320
+ More
UP000053105 UP000027135 UP000005203 UP000242457 UP000235965 UP000215335 UP000078540 UP000002358 UP000078492 UP000078541 UP000008820 UP000069940 UP000249989 UP000078542 UP000008237 UP000279307 UP000007062 UP000008792 UP000007266 UP000075902 UP000085678 UP000075883 UP000075900 UP000076407 UP000001070 UP000075920 UP000075882 UP000053825 UP000075903 UP000007801 UP000092553 UP000075840 UP000009192 UP000192221 UP000078200 UP000076420 UP000268350 UP000000304 UP000000803 UP000001292 UP000037069 UP000008711 UP000002282 UP000092443 UP000001819 UP000008744 UP000092460 UP000092461 UP000069272 UP000009046 UP000000673 UP000007798 UP000075884 UP000183832 UP000095301 UP000019118 UP000030742 UP000053097 UP000007819 UP000030746 UP000092445 UP000095300 UP000186698 UP000014760 UP000007755 UP000008143 UP000265180
UP000053105 UP000027135 UP000005203 UP000242457 UP000235965 UP000215335 UP000078540 UP000002358 UP000078492 UP000078541 UP000008820 UP000069940 UP000249989 UP000078542 UP000008237 UP000279307 UP000007062 UP000008792 UP000007266 UP000075902 UP000085678 UP000075883 UP000075900 UP000076407 UP000001070 UP000075920 UP000075882 UP000053825 UP000075903 UP000007801 UP000092553 UP000075840 UP000009192 UP000192221 UP000078200 UP000076420 UP000268350 UP000000304 UP000000803 UP000001292 UP000037069 UP000008711 UP000002282 UP000092443 UP000001819 UP000008744 UP000092460 UP000092461 UP000069272 UP000009046 UP000000673 UP000007798 UP000075884 UP000183832 UP000095301 UP000019118 UP000030742 UP000053097 UP000007819 UP000030746 UP000092445 UP000095300 UP000186698 UP000014760 UP000007755 UP000008143 UP000265180
Pfam
PF15469 Sec5
SUPFAM
SSF74788
SSF74788
ProteinModelPortal
H9JER6
A0A0L7LNG8
A0A2H1WXU1
A0A2W1BD04
A0A2A4JNG2
A0A194QK63
+ More
A0A194QIT2 A0A1E1W774 B0X770 A0A1Q3F808 A0A0N0BEZ7 A0A067QJH8 A0A088AF26 A0A2A3EV14 A0A2J7RFQ4 A0A232FNE9 A0A195BBN4 K7INY4 A0A151JCS4 A0A195FLH2 Q16KJ4 A0A182HB46 A0A1Y1ND64 A0A151IQH1 E2B9H0 A0A0C9PUE1 A0A3L8DDX5 A0A336LXL0 Q7Q6Z6 B4LJ46 D6X272 A0A182U4T4 A0A1S4FXK0 A0A1S3IN33 A0A182MUN4 A0A182RMJ1 A0A182XN22 B4J7M9 A0A182WKG8 A0A182LJA6 A0A0L7QYD5 A0A1B6MLS5 A0A182VH94 B3MI33 A0A1B6GXN5 A0A0M4EKU1 A0A182HWX9 B4KPL2 A0A1W4UAZ0 A0A1A9UY46 A0A2C9K343 A0A3B0J2W4 B4QD20 Q8MSY4 W8BZL6 B4HP05 A0A1W4UP77 A0A0L0CHZ7 B3NKA2 B4P4A1 A0A1A9Y0G8 Q291X2 B4GA52 A0A1B0B676 A0A0A1XN24 A0A1B0CB26 A0A0K8U7X5 A0A182FEH8 E0VIJ9 W5J5F2 B4MK00 A0A182NHQ4 A0A0K8U6Z8 A0A1J1J2L3 A0A1I8N657 A0A1I8N671 N6TX68 A0A2J7RFP7 A0A026WSA8 A0A1I8N667 A0A2S2NRF4 J9K7Q5 V4CEQ7 A0A0B6ZII1 A0A2H8TG56 A0A1A9ZY13 A0A1I8QD32 Q505L3 A0A1L8GCZ5 R7V5S5 F4WR83 A0A1I8QD70 Q4V9Y0 A0A1L8GJL8 A0A3B0IZ17 A0A3P9LE30 T1J0Z4 A0A1E1XI44
A0A194QIT2 A0A1E1W774 B0X770 A0A1Q3F808 A0A0N0BEZ7 A0A067QJH8 A0A088AF26 A0A2A3EV14 A0A2J7RFQ4 A0A232FNE9 A0A195BBN4 K7INY4 A0A151JCS4 A0A195FLH2 Q16KJ4 A0A182HB46 A0A1Y1ND64 A0A151IQH1 E2B9H0 A0A0C9PUE1 A0A3L8DDX5 A0A336LXL0 Q7Q6Z6 B4LJ46 D6X272 A0A182U4T4 A0A1S4FXK0 A0A1S3IN33 A0A182MUN4 A0A182RMJ1 A0A182XN22 B4J7M9 A0A182WKG8 A0A182LJA6 A0A0L7QYD5 A0A1B6MLS5 A0A182VH94 B3MI33 A0A1B6GXN5 A0A0M4EKU1 A0A182HWX9 B4KPL2 A0A1W4UAZ0 A0A1A9UY46 A0A2C9K343 A0A3B0J2W4 B4QD20 Q8MSY4 W8BZL6 B4HP05 A0A1W4UP77 A0A0L0CHZ7 B3NKA2 B4P4A1 A0A1A9Y0G8 Q291X2 B4GA52 A0A1B0B676 A0A0A1XN24 A0A1B0CB26 A0A0K8U7X5 A0A182FEH8 E0VIJ9 W5J5F2 B4MK00 A0A182NHQ4 A0A0K8U6Z8 A0A1J1J2L3 A0A1I8N657 A0A1I8N671 N6TX68 A0A2J7RFP7 A0A026WSA8 A0A1I8N667 A0A2S2NRF4 J9K7Q5 V4CEQ7 A0A0B6ZII1 A0A2H8TG56 A0A1A9ZY13 A0A1I8QD32 Q505L3 A0A1L8GCZ5 R7V5S5 F4WR83 A0A1I8QD70 Q4V9Y0 A0A1L8GJL8 A0A3B0IZ17 A0A3P9LE30 T1J0Z4 A0A1E1XI44
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus
Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
trans-Golgi network Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
Recycling endosome Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
trans-Golgi network Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
Recycling endosome Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
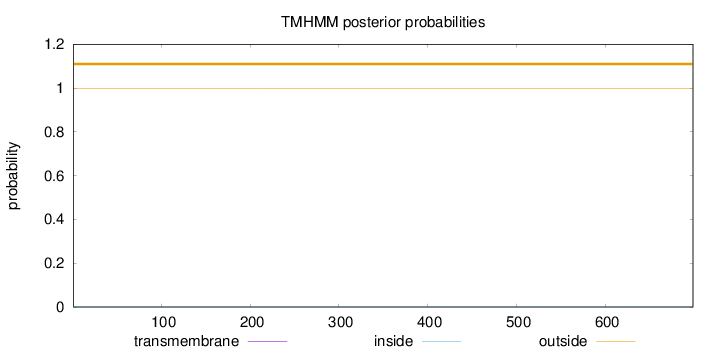
Length:
699
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0014
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00055
outside
1 - 699
Population Genetic Test Statistics
Pi
179.614427
Theta
222.982477
Tajima's D
-0.669202
CLR
308.500039
CSRT
0.202089895505225
Interpretation
Uncertain
