Gene
KWMTBOMO04909
Pre Gene Modal
BGIBMGA008069
Annotation
PREDICTED:_TBC1_domain_family_member_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.919 Nuclear Reliability : 1.661
Sequence
CDS
ATGGACCAGAATTCAGCGAAAACACCTAGTCTACCAAACAATGGTACGGAAGGGATTGATTTCGACCTGGAACAGCCGCTGTGGTGTTTGGAGGAGCTTCAGAAAGCGGCTATTGAGCATCGACTCCTGCGTCCACGGAGCCTGGCCTGGGCTATTATGCTGAACGCTTTGCCACCTCCGAATGCGGACATCGTTTCTCGCCTAAAGTGCCATAGGAACTTCTACGATGACTTGAAAGTGAAATTATCGATGGATCCGAGAGCCGTGATTGGGGATGACCCACTCTCTCAGAATGATAAGAGCGCTTGGCGACAGCACTTTTGCGACAACGAACTCAAAACTGTGATACTCCAAGACGTCGTGAGGACGTTTCCTGATGAGCTTTACTTTAGGGAGAAAGACGTTCAGGATCTGATGGTCCGAGTCCTCTTCTATTGGGCGCGTTCGCACTCGCACGTGGGCTACAGACAGGGCATGCACGAGATACTGGCTCCGCTACTCTTCGAGCTGTATTTGGATAGGAAGTTCGCGCCCCGGGCCATCAGTGACAAATTAAAAATTATACTCGACGAAGATTATTTGGAGCACGACAGCTACATGCTGTTCAACACCGTGATGCAGGGACTGGAGAGGTTTTACGTGACGGGCGACGTCGTGCCCTCGCCCTCCGGTAGGATGCCCTCTTCCAGGATGATCCACAATCCAAACGAAGTGGTGAGATACCTGGAGAAGGTCAAGGAGGACTACTTGTTCCCGCTGGACCCCGAGCTCGCCACGCACCTGACCAACTTCAACATTTCCTTGGAGCTCTTCGGAATGTGA
Protein
MDQNSAKTPSLPNNGTEGIDFDLEQPLWCLEELQKAAIEHRLLRPRSLAWAIMLNALPPPNADIVSRLKCHRNFYDDLKVKLSMDPRAVIGDDPLSQNDKSAWRQHFCDNELKTVILQDVVRTFPDELYFREKDVQDLMVRVLFYWARSHSHVGYRQGMHEILAPLLFELYLDRKFAPRAISDKLKIILDEDYLEHDSYMLFNTVMQGLERFYVTGDVVPSPSGRMPSSRMIHNPNEVVRYLEKVKEDYLFPLDPELATHLTNFNISLELFGM
Summary
Uniprot
A0A2W1B9Y5
A0A194QD23
A0A1E1WDQ2
A0A212F6L0
A0A1E1WK08
A0A1E1W7C2
+ More
J9JR80 A0A2S2R2X9 A0A0P4VI03 A0A023F190 A0A2H8TWH1 E0VZ09 A0A2S2P171 A0A224XBK3 A0A067RW36 A0A1B6HL30 A0A2R7VX54 A0A0V0G8C2 A0A2J7RE47 A0A2C9L1R7 A0A2C9L1K8 A0A224Z5N2 A0A131YT10 A0A224ZBW3 A0A2C9L1Q9 A0A1E1X3F0 A0A023G3B0 A0A0K8SS88 A0A0A9ZED7 A0A1S4FFS5 Q173B2 A0A0P4WL95 A0A1V9XZJ1 A0A0D2X3X9 A0A1E1XM41 W5JQF6 A0A2C9L1R4 R7U6Y5 A0A132A9H4 A0A2M4CMH5 A0A3B0KRL7 A0A1Q3F4H4 A0A1Q3F429 W8BMC7 A0A1Q3F4A3 B4IPE3 A0A3B0KHH7 A0A210PVT8 B4K9T6 A0A336MC51 A0A182FA64 Q299S6 B4G5N6 A0A2C9L1G2 A0A2C9L1V4 W8C6E8 A0A2R5LJS5 B3LX55 A0A293LF98 A0A034VTP3 A0A182UY02 Q7QDJ6 A0A182XK66 B4R1U5 A0A0K8UZC2 A0A293MZ68 A0A182NKV1 A0A182PZC0 A0A131YRW1 A0A182PS53 Q6PGX8 A0A1I8MJC2 A0A1I8MJA9 A0A182TVP1 A0A182IV38 A0A0A1XFL6 A0A182HJS2 A0A1Y9HF71 F1QXM3 A0A1W4UJX8 B3P168 B4PPY5 A0A084VZD4 B4HG96 B4M6D3 A0A182KWZ3 Q9VFX6 A0A1Y2CWE0 B1WC98 A0A182Y4I0 A0A2T7PV67
J9JR80 A0A2S2R2X9 A0A0P4VI03 A0A023F190 A0A2H8TWH1 E0VZ09 A0A2S2P171 A0A224XBK3 A0A067RW36 A0A1B6HL30 A0A2R7VX54 A0A0V0G8C2 A0A2J7RE47 A0A2C9L1R7 A0A2C9L1K8 A0A224Z5N2 A0A131YT10 A0A224ZBW3 A0A2C9L1Q9 A0A1E1X3F0 A0A023G3B0 A0A0K8SS88 A0A0A9ZED7 A0A1S4FFS5 Q173B2 A0A0P4WL95 A0A1V9XZJ1 A0A0D2X3X9 A0A1E1XM41 W5JQF6 A0A2C9L1R4 R7U6Y5 A0A132A9H4 A0A2M4CMH5 A0A3B0KRL7 A0A1Q3F4H4 A0A1Q3F429 W8BMC7 A0A1Q3F4A3 B4IPE3 A0A3B0KHH7 A0A210PVT8 B4K9T6 A0A336MC51 A0A182FA64 Q299S6 B4G5N6 A0A2C9L1G2 A0A2C9L1V4 W8C6E8 A0A2R5LJS5 B3LX55 A0A293LF98 A0A034VTP3 A0A182UY02 Q7QDJ6 A0A182XK66 B4R1U5 A0A0K8UZC2 A0A293MZ68 A0A182NKV1 A0A182PZC0 A0A131YRW1 A0A182PS53 Q6PGX8 A0A1I8MJC2 A0A1I8MJA9 A0A182TVP1 A0A182IV38 A0A0A1XFL6 A0A182HJS2 A0A1Y9HF71 F1QXM3 A0A1W4UJX8 B3P168 B4PPY5 A0A084VZD4 B4HG96 B4M6D3 A0A182KWZ3 Q9VFX6 A0A1Y2CWE0 B1WC98 A0A182Y4I0 A0A2T7PV67
Pubmed
28756777
26354079
22118469
27129103
25474469
20566863
+ More
24845553 15562597 28797301 26830274 28503490 25401762 26823975 17510324 28327890 29209593 20920257 23761445 23254933 26555130 24495485 17994087 28812685 15632085 25348373 12364791 14747013 17210077 25315136 25830018 23594743 17550304 24438588 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15489334 25244985
24845553 15562597 28797301 26830274 28503490 25401762 26823975 17510324 28327890 29209593 20920257 23761445 23254933 26555130 24495485 17994087 28812685 15632085 25348373 12364791 14747013 17210077 25315136 25830018 23594743 17550304 24438588 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15489334 25244985
EMBL
KZ150199
PZC72292.1
KQ459185
KPJ03362.1
GDQN01005941
JAT85113.1
+ More
AGBW02009996 OWR49375.1 GDQN01003739 JAT87315.1 GDQN01008164 JAT82890.1 ABLF02029495 GGMS01015153 MBY84356.1 GDKW01003221 JAI53374.1 GBBI01003948 JAC14764.1 GFXV01006791 MBW18596.1 DS235848 EEB18615.1 GGMR01010536 MBY23155.1 GFTR01006629 JAW09797.1 KK852427 KDR24084.1 GECU01032328 JAS75378.1 KK854144 PTY12083.1 GECL01001737 JAP04387.1 NEVH01005277 PNF39109.1 GFPF01013409 MAA24555.1 GEDV01007281 JAP81276.1 GFPF01013408 MAA24554.1 GFAC01005411 JAT93777.1 GBBM01006732 JAC28686.1 GBRD01009788 GBRD01009787 JAG56036.1 GBHO01003444 GBHO01000184 GDHC01013088 GDHC01008572 JAG40160.1 JAG43420.1 JAQ05541.1 JAQ10057.1 CH477428 EAT41116.1 GDRN01019106 JAI67819.1 MNPL01001705 OQR78891.1 KE346368 KJE95089.1 GFAA01003106 JAU00329.1 ADMH02000806 ETN64974.1 AMQN01008986 KB304341 ELU02125.1 JXLN01011622 KPM07519.1 GGFL01002348 MBW66526.1 OUUW01000013 SPP87891.1 GFDL01012579 JAV22466.1 GFDL01012731 JAV22314.1 GAMC01004305 JAC02251.1 GFDL01012696 JAV22349.1 CH680252 EDW53727.1 SPP87890.1 NEDP02005459 OWF40600.1 CH933806 EDW14561.2 UFQT01000635 SSX25967.1 CM000070 EAL27625.2 CH479179 EDW24902.1 GAMC01004304 JAC02252.1 GGLE01005549 MBY09675.1 CH902617 EDV41655.1 GFWV01002444 MAA27174.1 GAKP01013465 JAC45487.1 AAAB01008851 EAA07272.5 EGK96624.1 CM000364 EDX13097.1 GDHF01020305 JAI32009.1 GFWV01021375 MAA46103.1 AXCN02000306 GEDV01007282 JAP81275.1 BC056792 AAH56792.1 GBXI01005014 JAD09278.1 APCN01002207 BX927400 CH954181 EDV49257.1 CM000160 EDW97212.1 ATLV01018745 KE525248 KFB43328.1 CH480815 EDW42344.1 CH940652 EDW59209.2 AE014297 BT023950 AAF54919.1 ABB36454.1 MCGO01000005 ORY51340.1 BC162056 AAI62056.1 PZQS01000002 PVD37311.1
AGBW02009996 OWR49375.1 GDQN01003739 JAT87315.1 GDQN01008164 JAT82890.1 ABLF02029495 GGMS01015153 MBY84356.1 GDKW01003221 JAI53374.1 GBBI01003948 JAC14764.1 GFXV01006791 MBW18596.1 DS235848 EEB18615.1 GGMR01010536 MBY23155.1 GFTR01006629 JAW09797.1 KK852427 KDR24084.1 GECU01032328 JAS75378.1 KK854144 PTY12083.1 GECL01001737 JAP04387.1 NEVH01005277 PNF39109.1 GFPF01013409 MAA24555.1 GEDV01007281 JAP81276.1 GFPF01013408 MAA24554.1 GFAC01005411 JAT93777.1 GBBM01006732 JAC28686.1 GBRD01009788 GBRD01009787 JAG56036.1 GBHO01003444 GBHO01000184 GDHC01013088 GDHC01008572 JAG40160.1 JAG43420.1 JAQ05541.1 JAQ10057.1 CH477428 EAT41116.1 GDRN01019106 JAI67819.1 MNPL01001705 OQR78891.1 KE346368 KJE95089.1 GFAA01003106 JAU00329.1 ADMH02000806 ETN64974.1 AMQN01008986 KB304341 ELU02125.1 JXLN01011622 KPM07519.1 GGFL01002348 MBW66526.1 OUUW01000013 SPP87891.1 GFDL01012579 JAV22466.1 GFDL01012731 JAV22314.1 GAMC01004305 JAC02251.1 GFDL01012696 JAV22349.1 CH680252 EDW53727.1 SPP87890.1 NEDP02005459 OWF40600.1 CH933806 EDW14561.2 UFQT01000635 SSX25967.1 CM000070 EAL27625.2 CH479179 EDW24902.1 GAMC01004304 JAC02252.1 GGLE01005549 MBY09675.1 CH902617 EDV41655.1 GFWV01002444 MAA27174.1 GAKP01013465 JAC45487.1 AAAB01008851 EAA07272.5 EGK96624.1 CM000364 EDX13097.1 GDHF01020305 JAI32009.1 GFWV01021375 MAA46103.1 AXCN02000306 GEDV01007282 JAP81275.1 BC056792 AAH56792.1 GBXI01005014 JAD09278.1 APCN01002207 BX927400 CH954181 EDV49257.1 CM000160 EDW97212.1 ATLV01018745 KE525248 KFB43328.1 CH480815 EDW42344.1 CH940652 EDW59209.2 AE014297 BT023950 AAF54919.1 ABB36454.1 MCGO01000005 ORY51340.1 BC162056 AAI62056.1 PZQS01000002 PVD37311.1
Proteomes
UP000053268
UP000007151
UP000007819
UP000009046
UP000027135
UP000235965
+ More
UP000076420 UP000008820 UP000192247 UP000008743 UP000000673 UP000014760 UP000268350 UP000001292 UP000242188 UP000009192 UP000069272 UP000001819 UP000008744 UP000007801 UP000075903 UP000007062 UP000076407 UP000000304 UP000075884 UP000075886 UP000075885 UP000095301 UP000075902 UP000075880 UP000075840 UP000075900 UP000000437 UP000192221 UP000008711 UP000002282 UP000030765 UP000008792 UP000075882 UP000000803 UP000193642 UP000076408 UP000245119
UP000076420 UP000008820 UP000192247 UP000008743 UP000000673 UP000014760 UP000268350 UP000001292 UP000242188 UP000009192 UP000069272 UP000001819 UP000008744 UP000007801 UP000075903 UP000007062 UP000076407 UP000000304 UP000075884 UP000075886 UP000075885 UP000095301 UP000075902 UP000075880 UP000075840 UP000075900 UP000000437 UP000192221 UP000008711 UP000002282 UP000030765 UP000008792 UP000075882 UP000000803 UP000193642 UP000076408 UP000245119
PRIDE
Interpro
SUPFAM
SSF47923
SSF47923
Gene 3D
ProteinModelPortal
A0A2W1B9Y5
A0A194QD23
A0A1E1WDQ2
A0A212F6L0
A0A1E1WK08
A0A1E1W7C2
+ More
J9JR80 A0A2S2R2X9 A0A0P4VI03 A0A023F190 A0A2H8TWH1 E0VZ09 A0A2S2P171 A0A224XBK3 A0A067RW36 A0A1B6HL30 A0A2R7VX54 A0A0V0G8C2 A0A2J7RE47 A0A2C9L1R7 A0A2C9L1K8 A0A224Z5N2 A0A131YT10 A0A224ZBW3 A0A2C9L1Q9 A0A1E1X3F0 A0A023G3B0 A0A0K8SS88 A0A0A9ZED7 A0A1S4FFS5 Q173B2 A0A0P4WL95 A0A1V9XZJ1 A0A0D2X3X9 A0A1E1XM41 W5JQF6 A0A2C9L1R4 R7U6Y5 A0A132A9H4 A0A2M4CMH5 A0A3B0KRL7 A0A1Q3F4H4 A0A1Q3F429 W8BMC7 A0A1Q3F4A3 B4IPE3 A0A3B0KHH7 A0A210PVT8 B4K9T6 A0A336MC51 A0A182FA64 Q299S6 B4G5N6 A0A2C9L1G2 A0A2C9L1V4 W8C6E8 A0A2R5LJS5 B3LX55 A0A293LF98 A0A034VTP3 A0A182UY02 Q7QDJ6 A0A182XK66 B4R1U5 A0A0K8UZC2 A0A293MZ68 A0A182NKV1 A0A182PZC0 A0A131YRW1 A0A182PS53 Q6PGX8 A0A1I8MJC2 A0A1I8MJA9 A0A182TVP1 A0A182IV38 A0A0A1XFL6 A0A182HJS2 A0A1Y9HF71 F1QXM3 A0A1W4UJX8 B3P168 B4PPY5 A0A084VZD4 B4HG96 B4M6D3 A0A182KWZ3 Q9VFX6 A0A1Y2CWE0 B1WC98 A0A182Y4I0 A0A2T7PV67
J9JR80 A0A2S2R2X9 A0A0P4VI03 A0A023F190 A0A2H8TWH1 E0VZ09 A0A2S2P171 A0A224XBK3 A0A067RW36 A0A1B6HL30 A0A2R7VX54 A0A0V0G8C2 A0A2J7RE47 A0A2C9L1R7 A0A2C9L1K8 A0A224Z5N2 A0A131YT10 A0A224ZBW3 A0A2C9L1Q9 A0A1E1X3F0 A0A023G3B0 A0A0K8SS88 A0A0A9ZED7 A0A1S4FFS5 Q173B2 A0A0P4WL95 A0A1V9XZJ1 A0A0D2X3X9 A0A1E1XM41 W5JQF6 A0A2C9L1R4 R7U6Y5 A0A132A9H4 A0A2M4CMH5 A0A3B0KRL7 A0A1Q3F4H4 A0A1Q3F429 W8BMC7 A0A1Q3F4A3 B4IPE3 A0A3B0KHH7 A0A210PVT8 B4K9T6 A0A336MC51 A0A182FA64 Q299S6 B4G5N6 A0A2C9L1G2 A0A2C9L1V4 W8C6E8 A0A2R5LJS5 B3LX55 A0A293LF98 A0A034VTP3 A0A182UY02 Q7QDJ6 A0A182XK66 B4R1U5 A0A0K8UZC2 A0A293MZ68 A0A182NKV1 A0A182PZC0 A0A131YRW1 A0A182PS53 Q6PGX8 A0A1I8MJC2 A0A1I8MJA9 A0A182TVP1 A0A182IV38 A0A0A1XFL6 A0A182HJS2 A0A1Y9HF71 F1QXM3 A0A1W4UJX8 B3P168 B4PPY5 A0A084VZD4 B4HG96 B4M6D3 A0A182KWZ3 Q9VFX6 A0A1Y2CWE0 B1WC98 A0A182Y4I0 A0A2T7PV67
PDB
2G77
E-value=1.40531e-05,
Score=114
Ontologies
GO
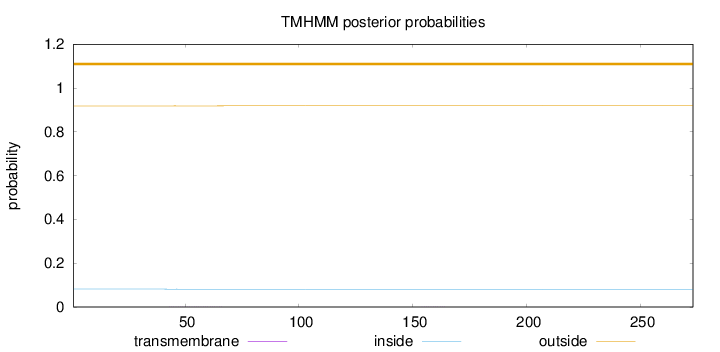
Topology
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0214599999999999
Exp number, first 60 AAs:
0.01334
Total prob of N-in:
0.08166
outside
1 - 273
Population Genetic Test Statistics
Pi
2.445897
Theta
54.57378
Tajima's D
-1.998556
CLR
2.421003
CSRT
0.0135993200339983
Interpretation
Possibly Positive selection
