Gene
KWMTBOMO04907
Pre Gene Modal
BGIBMGA008067
Annotation
PREDICTED:_transmembrane_protein_181_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.854
Sequence
CDS
ATGGCTTATAACGATTTCATTGACATCAAACAACTGCCCGTTCCTAGACTGAAAAAAAGCGTTGAAAGTGAGCTTGTAAAGGTGTCTGATATAAAAGTCATCAAAGTGGTGAAAGTTGAAGATAATAAGGTAAAATTTTGTTACAAAACATCCTCCGTTGATGATTTCAAAGAACTTAACCTGGGATCTAAAAGAGCCAGTGCAAGGAACCAGAGGACTGAAGAATTGCAACATCTGTATAACCAAAACGACTTGTTCAGTGAATTCAACAAATATATAGCTCCGGCATATCACCATGACAGGTGTGAAAGGTCGGTTCAAATGAGGATATATTCAATGCACAAAGGGGAATTCGTGATGGTTTTTATAGCATTTTTTGCTTGCTTCGGTCTCGGAGTTTTCATAGGACTGGCAGGGCCCTCGCCTACTCTGACTACGTCAATAACAGCGAGCAGTGTGCTGGATAACGGCAGCATGATATACCAGGGACCCTTCCACCTCCACAGCCCAGCCCTGGGCACCAGAGTGCAGCAGCTGTGGCTGTTCGCCGAAATACTCACGGAGAATAATGATGAGGAAATATTTGACAAGAGCTTCCAGATTAGTATATCTATAGACGGCGTTCGCGCCGACCATTCCACCATCAACTTACTCCCAGAAAACGAATCCACGAATACGACACAACATCTGAAATGCCACAAGCAGCTGTGCGAGGAAGTTATGGTGTTGCACCTGGCCTCGCTCGAGTACACTCACTACATACTGAACATTCGCTTCTACGGGCTCGGGGAGTTCCACAAACGTTACTACATGAGGGAGATTGTGTTCTATTTTAAAATGTACAACCCAGCGTTCACGCAAATGGAGACTTGGTTCAGATTTATTTTCCTGTTGGCCACATTTGTTGTTACGTGCTGGTTCACGCACTCCCTCAGGAAGTATCCGTCCCACGACTGGGCGATCGAGCAGCGCTGGCTGTCAGTGATGCTGCCGATGCTGCTCCTCTACAACGATCCGCTGTTCCCGCTGCGGCTGGTGTCAGCGAGCTGCTTCGCGCCGCTGATGGACTCTGTGTTCCAGGCGACCTTCTTCGCGGGCCTCATGCTGTCCTGGCTGGCACAATATCACGGGCTGCGACAGAACGACCGCGGCTTCGTGTCGTTCTACCTGTTCAAGCTGGTGGTGGTGGGCTCGGTGTGGGCGCCGGCGATGGTCGTCACCGTCTGGCAGAAATATTACGCGTATTACGATCCCACCTTCAACTACATGATGTCGCCCAACTATCCGATCGTAAAGACGATGTTCTTCACGGGCCTCGCCGTGTACTTCCTGTACCTACTCATACTCGTGGTGAAGGCCTATAGTGATTTGAGGAACATGCCGTTTTTTGACGTGCGGCTGCGCTGCGTGTCTGCGGGCGCGGGCGCGGCGGCGGCGCTGGCGGGCGCGGTGTGCGCGCGGGGGTGGGGCGGCGCGGCGCTGCAGGACCACTGGGCGGCGCGGCACCCGCACGACACGTCCGCGCCCTTCATGGCGCTCTACTCGCTCTTCAACTTCGACCTCTACGCGCTCGCCTACCTGTACTCGCCGCCCACGGTCGAGCCCCACGAAACGTCCATAACAAAAGACAATCCCGCCTTCTCGATGATCAACGATTCGGACGAGGACGTCATCTATGGCTCGGACGAAGACAGCCGGCGGCCCCTCAACTCGCACCACCGAGTCAGCACAGAAGACATATGA
Protein
MAYNDFIDIKQLPVPRLKKSVESELVKVSDIKVIKVVKVEDNKVKFCYKTSSVDDFKELNLGSKRASARNQRTEELQHLYNQNDLFSEFNKYIAPAYHHDRCERSVQMRIYSMHKGEFVMVFIAFFACFGLGVFIGLAGPSPTLTTSITASSVLDNGSMIYQGPFHLHSPALGTRVQQLWLFAEILTENNDEEIFDKSFQISISIDGVRADHSTINLLPENESTNTTQHLKCHKQLCEEVMVLHLASLEYTHYILNIRFYGLGEFHKRYYMREIVFYFKMYNPAFTQMETWFRFIFLLATFVVTCWFTHSLRKYPSHDWAIEQRWLSVMLPMLLLYNDPLFPLRLVSASCFAPLMDSVFQATFFAGLMLSWLAQYHGLRQNDRGFVSFYLFKLVVVGSVWAPAMVVTVWQKYYAYYDPTFNYMMSPNYPIVKTMFFTGLAVYFLYLLILVVKAYSDLRNMPFFDVRLRCVSAGAGAAAALAGAVCARGWGGAALQDHWAARHPHDTSAPFMALYSLFNFDLYALAYLYSPPTVEPHETSITKDNPAFSMINDSDEDVIYGSDEDSRRPLNSHHRVSTEDI
Summary
Uniprot
H9JEX0
A0A3S2TEZ0
A0A194QR08
A0A194QD68
A0A2A4JF46
A0A2W1BF27
+ More
A0A2H1WZI8 K7J004 A0A1W4X1X9 A0A1S4F711 T1I0C1 Q17DF1 A0A182G299 A0A0C9RL15 A0A2J7R175 D6X3S7 A0A067REA9 E2BAS9 W8C2S4 A0A195CH58 E9IWU5 A0A0K8UEN7 A0A195F2A0 F4X6R5 E2A2K1 W8BMX4 A0A195B1Y7 A0A158NFU4 A0A195E7K4 A0A026X1C8 A0A0K8ULX4 A0A336M118 E9FZ18 A0A1I8N4L8 B0X9A5 B4JRL7 A0A182NQA4 A0A164QU79 A0A2A3EFV4 A0A1I8Q930 B4M0N1 A0A0L7RDW7 A0A1B0FR64 A0A0L0CHU4 B4K6L2 A0A182QT09 A0A182JCN4 A0A0P5GF15 A0A1B0GLK6 A0A0M4ENX6 A0A0P4XFW7 A0A084VJE0 A0A1B0C2U9 A0A0P5GX83 A0A1W4VQC6 B4HM82 Q297T8 A0A0R1E940 B4G310 A0A151XC78 B4QZD2 Q9VD53 B4NHR4 B4PLJ7 B3P7P0 A0A182M4I0 A0A182W463 A0A182KGU3 B3LVL2 A0A3B0KBV1 A0A182RH92 A0A088AN49 A0A182Y557 A0A1A9UZT2 A0A182FMC6 A0A182I9K7 A0A182TF24 A0A182USA3 Q7QE84 A0A2S2N7F8 A0A2P6L0N3 A0A2H8TR42 A0A182P6T6 A0A232F6C4 A0A0M8ZYK8 A0A182X5D0 Q4V5Q6 N6TN99 Q4V642 J9K1U2 A0A0R3NP09 A0A226F2D7 A0A1W4XCM1 T1J7Y3 A0A1A9ZZP9 A0A0P5BCI6 A0A1B6LXN5 A0A2J7R171 A0A0P5JLE2 A0A1S3JDT1
A0A2H1WZI8 K7J004 A0A1W4X1X9 A0A1S4F711 T1I0C1 Q17DF1 A0A182G299 A0A0C9RL15 A0A2J7R175 D6X3S7 A0A067REA9 E2BAS9 W8C2S4 A0A195CH58 E9IWU5 A0A0K8UEN7 A0A195F2A0 F4X6R5 E2A2K1 W8BMX4 A0A195B1Y7 A0A158NFU4 A0A195E7K4 A0A026X1C8 A0A0K8ULX4 A0A336M118 E9FZ18 A0A1I8N4L8 B0X9A5 B4JRL7 A0A182NQA4 A0A164QU79 A0A2A3EFV4 A0A1I8Q930 B4M0N1 A0A0L7RDW7 A0A1B0FR64 A0A0L0CHU4 B4K6L2 A0A182QT09 A0A182JCN4 A0A0P5GF15 A0A1B0GLK6 A0A0M4ENX6 A0A0P4XFW7 A0A084VJE0 A0A1B0C2U9 A0A0P5GX83 A0A1W4VQC6 B4HM82 Q297T8 A0A0R1E940 B4G310 A0A151XC78 B4QZD2 Q9VD53 B4NHR4 B4PLJ7 B3P7P0 A0A182M4I0 A0A182W463 A0A182KGU3 B3LVL2 A0A3B0KBV1 A0A182RH92 A0A088AN49 A0A182Y557 A0A1A9UZT2 A0A182FMC6 A0A182I9K7 A0A182TF24 A0A182USA3 Q7QE84 A0A2S2N7F8 A0A2P6L0N3 A0A2H8TR42 A0A182P6T6 A0A232F6C4 A0A0M8ZYK8 A0A182X5D0 Q4V5Q6 N6TN99 Q4V642 J9K1U2 A0A0R3NP09 A0A226F2D7 A0A1W4XCM1 T1J7Y3 A0A1A9ZZP9 A0A0P5BCI6 A0A1B6LXN5 A0A2J7R171 A0A0P5JLE2 A0A1S3JDT1
Pubmed
19121390
26354079
28756777
20075255
17510324
26483478
+ More
18362917 19820115 24845553 20798317 24495485 21282665 21719571 21347285 24508170 30249741 21292972 25315136 17994087 26108605 24438588 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 12364791 14747013 17210077 28648823 23537049
18362917 19820115 24845553 20798317 24495485 21282665 21719571 21347285 24508170 30249741 21292972 25315136 17994087 26108605 24438588 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 12364791 14747013 17210077 28648823 23537049
EMBL
BABH01023421
RSAL01000211
RVE44191.1
KQ461198
KPJ05966.1
KQ459185
+ More
KPJ03364.1 NWSH01001607 PCG70715.1 KZ150199 PZC72294.1 ODYU01012276 SOQ58495.1 ACPB03018331 CH477296 EAT44386.1 JXUM01039735 JXUM01039736 JXUM01039737 JXUM01039738 KQ561216 KXJ79321.1 GBYB01008905 GBYB01008909 JAG78672.1 JAG78676.1 NEVH01008213 PNF34583.1 KQ971326 EEZ97376.1 KK852517 KDR22181.1 GL446831 EFN87214.1 GAMC01006414 JAC00142.1 KQ977754 KYN00071.1 GL766614 EFZ14953.1 GDHF01027200 JAI25114.1 KQ981864 KYN34302.1 GL888818 EGI57847.1 GL436083 EFN72339.1 GAMC01006413 GAMC01006412 JAC00143.1 KQ976662 KYM78491.1 ADTU01014740 ADTU01014741 KQ979568 KYN20832.1 KK107063 QOIP01000005 EZA61174.1 RLU22352.1 GDHF01031295 GDHF01024964 JAI21019.1 JAI27350.1 UFQT01000302 SSX23021.1 GL732527 EFX87631.1 DS232529 EDS43037.1 CH916373 EDV94407.1 LRGB01002371 KZS08062.1 KZ288269 PBC30036.1 CH940650 EDW67323.1 KQ414613 KOC68926.1 CCAG010008016 JRES01000454 KNC31059.1 CH933806 EDW16312.1 AXCN02001293 GDIQ01242360 JAK09365.1 AJWK01035621 CP012526 ALC47402.1 GDIP01243706 GDIP01203387 JAI79695.1 ATLV01013544 KE524867 KFB38084.1 JXJN01024707 GDIQ01257570 JAJ94154.1 CH480815 EDW43130.1 CM000070 EAL28117.1 CM000160 KRK04034.1 CH479179 EDW24205.1 KQ982314 KYQ57964.1 CM000364 EDX13868.1 AE014297 BT133377 AAF55948.1 AFH36362.1 CH964272 EDW84674.1 EDW97946.2 CH954182 EDV54201.1 AXCM01001166 CH902617 EDV42582.1 OUUW01000005 SPP81058.1 APCN01000990 AAAB01008847 EAA06895.3 GGMR01000097 MBY12716.1 MWRG01002896 PRD32116.1 GFXV01004820 MBW16625.1 NNAY01000891 OXU26040.1 KQ435824 KOX72219.1 BT022600 AAY55016.1 APGK01017227 KB739998 ENN81969.1 BT022464 AAY54880.1 ABLF02024067 ABLF02024071 KRT00470.1 LNIX01000001 OXA63106.1 JH431944 GDIP01186426 JAJ36976.1 GEBQ01011587 JAT28390.1 PNF34581.1 GDIQ01203261 JAK48464.1
KPJ03364.1 NWSH01001607 PCG70715.1 KZ150199 PZC72294.1 ODYU01012276 SOQ58495.1 ACPB03018331 CH477296 EAT44386.1 JXUM01039735 JXUM01039736 JXUM01039737 JXUM01039738 KQ561216 KXJ79321.1 GBYB01008905 GBYB01008909 JAG78672.1 JAG78676.1 NEVH01008213 PNF34583.1 KQ971326 EEZ97376.1 KK852517 KDR22181.1 GL446831 EFN87214.1 GAMC01006414 JAC00142.1 KQ977754 KYN00071.1 GL766614 EFZ14953.1 GDHF01027200 JAI25114.1 KQ981864 KYN34302.1 GL888818 EGI57847.1 GL436083 EFN72339.1 GAMC01006413 GAMC01006412 JAC00143.1 KQ976662 KYM78491.1 ADTU01014740 ADTU01014741 KQ979568 KYN20832.1 KK107063 QOIP01000005 EZA61174.1 RLU22352.1 GDHF01031295 GDHF01024964 JAI21019.1 JAI27350.1 UFQT01000302 SSX23021.1 GL732527 EFX87631.1 DS232529 EDS43037.1 CH916373 EDV94407.1 LRGB01002371 KZS08062.1 KZ288269 PBC30036.1 CH940650 EDW67323.1 KQ414613 KOC68926.1 CCAG010008016 JRES01000454 KNC31059.1 CH933806 EDW16312.1 AXCN02001293 GDIQ01242360 JAK09365.1 AJWK01035621 CP012526 ALC47402.1 GDIP01243706 GDIP01203387 JAI79695.1 ATLV01013544 KE524867 KFB38084.1 JXJN01024707 GDIQ01257570 JAJ94154.1 CH480815 EDW43130.1 CM000070 EAL28117.1 CM000160 KRK04034.1 CH479179 EDW24205.1 KQ982314 KYQ57964.1 CM000364 EDX13868.1 AE014297 BT133377 AAF55948.1 AFH36362.1 CH964272 EDW84674.1 EDW97946.2 CH954182 EDV54201.1 AXCM01001166 CH902617 EDV42582.1 OUUW01000005 SPP81058.1 APCN01000990 AAAB01008847 EAA06895.3 GGMR01000097 MBY12716.1 MWRG01002896 PRD32116.1 GFXV01004820 MBW16625.1 NNAY01000891 OXU26040.1 KQ435824 KOX72219.1 BT022600 AAY55016.1 APGK01017227 KB739998 ENN81969.1 BT022464 AAY54880.1 ABLF02024067 ABLF02024071 KRT00470.1 LNIX01000001 OXA63106.1 JH431944 GDIP01186426 JAJ36976.1 GEBQ01011587 JAT28390.1 PNF34581.1 GDIQ01203261 JAK48464.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000002358
+ More
UP000192223 UP000015103 UP000008820 UP000069940 UP000249989 UP000235965 UP000007266 UP000027135 UP000008237 UP000078542 UP000078541 UP000007755 UP000000311 UP000078540 UP000005205 UP000078492 UP000053097 UP000279307 UP000000305 UP000095301 UP000002320 UP000001070 UP000075884 UP000076858 UP000242457 UP000095300 UP000008792 UP000053825 UP000092444 UP000037069 UP000009192 UP000075886 UP000075880 UP000092461 UP000092553 UP000030765 UP000092460 UP000192221 UP000001292 UP000001819 UP000002282 UP000008744 UP000075809 UP000000304 UP000000803 UP000007798 UP000008711 UP000075883 UP000075920 UP000075881 UP000007801 UP000268350 UP000075900 UP000005203 UP000076408 UP000078200 UP000069272 UP000075840 UP000075902 UP000075903 UP000007062 UP000075885 UP000215335 UP000053105 UP000076407 UP000019118 UP000007819 UP000198287 UP000092445 UP000085678
UP000192223 UP000015103 UP000008820 UP000069940 UP000249989 UP000235965 UP000007266 UP000027135 UP000008237 UP000078542 UP000078541 UP000007755 UP000000311 UP000078540 UP000005205 UP000078492 UP000053097 UP000279307 UP000000305 UP000095301 UP000002320 UP000001070 UP000075884 UP000076858 UP000242457 UP000095300 UP000008792 UP000053825 UP000092444 UP000037069 UP000009192 UP000075886 UP000075880 UP000092461 UP000092553 UP000030765 UP000092460 UP000192221 UP000001292 UP000001819 UP000002282 UP000008744 UP000075809 UP000000304 UP000000803 UP000007798 UP000008711 UP000075883 UP000075920 UP000075881 UP000007801 UP000268350 UP000075900 UP000005203 UP000076408 UP000078200 UP000069272 UP000075840 UP000075902 UP000075903 UP000007062 UP000075885 UP000215335 UP000053105 UP000076407 UP000019118 UP000007819 UP000198287 UP000092445 UP000085678
Interpro
IPR040416
TMEM181
+ More
IPR033581 APN2
IPR024571 ERAP1-like_C_dom
IPR014782 Peptidase_M1_dom
IPR034016 M1_APN-typ
IPR001930 Peptidase_M1
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR032847 PRPF17
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR020472 G-protein_beta_WD-40_rep
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR017893 DBB_domain
IPR035979 RBD_domain_sf
IPR034392 TatSF1-like_RRM1
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR003954 RRM_dom_euk
IPR033581 APN2
IPR024571 ERAP1-like_C_dom
IPR014782 Peptidase_M1_dom
IPR034016 M1_APN-typ
IPR001930 Peptidase_M1
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR032847 PRPF17
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR020472 G-protein_beta_WD-40_rep
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR017893 DBB_domain
IPR035979 RBD_domain_sf
IPR034392 TatSF1-like_RRM1
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR003954 RRM_dom_euk
Gene 3D
ProteinModelPortal
H9JEX0
A0A3S2TEZ0
A0A194QR08
A0A194QD68
A0A2A4JF46
A0A2W1BF27
+ More
A0A2H1WZI8 K7J004 A0A1W4X1X9 A0A1S4F711 T1I0C1 Q17DF1 A0A182G299 A0A0C9RL15 A0A2J7R175 D6X3S7 A0A067REA9 E2BAS9 W8C2S4 A0A195CH58 E9IWU5 A0A0K8UEN7 A0A195F2A0 F4X6R5 E2A2K1 W8BMX4 A0A195B1Y7 A0A158NFU4 A0A195E7K4 A0A026X1C8 A0A0K8ULX4 A0A336M118 E9FZ18 A0A1I8N4L8 B0X9A5 B4JRL7 A0A182NQA4 A0A164QU79 A0A2A3EFV4 A0A1I8Q930 B4M0N1 A0A0L7RDW7 A0A1B0FR64 A0A0L0CHU4 B4K6L2 A0A182QT09 A0A182JCN4 A0A0P5GF15 A0A1B0GLK6 A0A0M4ENX6 A0A0P4XFW7 A0A084VJE0 A0A1B0C2U9 A0A0P5GX83 A0A1W4VQC6 B4HM82 Q297T8 A0A0R1E940 B4G310 A0A151XC78 B4QZD2 Q9VD53 B4NHR4 B4PLJ7 B3P7P0 A0A182M4I0 A0A182W463 A0A182KGU3 B3LVL2 A0A3B0KBV1 A0A182RH92 A0A088AN49 A0A182Y557 A0A1A9UZT2 A0A182FMC6 A0A182I9K7 A0A182TF24 A0A182USA3 Q7QE84 A0A2S2N7F8 A0A2P6L0N3 A0A2H8TR42 A0A182P6T6 A0A232F6C4 A0A0M8ZYK8 A0A182X5D0 Q4V5Q6 N6TN99 Q4V642 J9K1U2 A0A0R3NP09 A0A226F2D7 A0A1W4XCM1 T1J7Y3 A0A1A9ZZP9 A0A0P5BCI6 A0A1B6LXN5 A0A2J7R171 A0A0P5JLE2 A0A1S3JDT1
A0A2H1WZI8 K7J004 A0A1W4X1X9 A0A1S4F711 T1I0C1 Q17DF1 A0A182G299 A0A0C9RL15 A0A2J7R175 D6X3S7 A0A067REA9 E2BAS9 W8C2S4 A0A195CH58 E9IWU5 A0A0K8UEN7 A0A195F2A0 F4X6R5 E2A2K1 W8BMX4 A0A195B1Y7 A0A158NFU4 A0A195E7K4 A0A026X1C8 A0A0K8ULX4 A0A336M118 E9FZ18 A0A1I8N4L8 B0X9A5 B4JRL7 A0A182NQA4 A0A164QU79 A0A2A3EFV4 A0A1I8Q930 B4M0N1 A0A0L7RDW7 A0A1B0FR64 A0A0L0CHU4 B4K6L2 A0A182QT09 A0A182JCN4 A0A0P5GF15 A0A1B0GLK6 A0A0M4ENX6 A0A0P4XFW7 A0A084VJE0 A0A1B0C2U9 A0A0P5GX83 A0A1W4VQC6 B4HM82 Q297T8 A0A0R1E940 B4G310 A0A151XC78 B4QZD2 Q9VD53 B4NHR4 B4PLJ7 B3P7P0 A0A182M4I0 A0A182W463 A0A182KGU3 B3LVL2 A0A3B0KBV1 A0A182RH92 A0A088AN49 A0A182Y557 A0A1A9UZT2 A0A182FMC6 A0A182I9K7 A0A182TF24 A0A182USA3 Q7QE84 A0A2S2N7F8 A0A2P6L0N3 A0A2H8TR42 A0A182P6T6 A0A232F6C4 A0A0M8ZYK8 A0A182X5D0 Q4V5Q6 N6TN99 Q4V642 J9K1U2 A0A0R3NP09 A0A226F2D7 A0A1W4XCM1 T1J7Y3 A0A1A9ZZP9 A0A0P5BCI6 A0A1B6LXN5 A0A2J7R171 A0A0P5JLE2 A0A1S3JDT1
Ontologies
GO
PANTHER
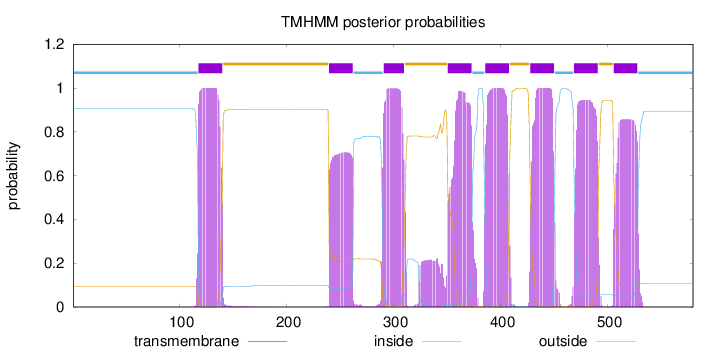
Topology
Length:
580
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
169.70096
Exp number, first 60 AAs:
0
Total prob of N-in:
0.90556
inside
1 - 117
TMhelix
118 - 140
outside
141 - 239
TMhelix
240 - 262
inside
263 - 290
TMhelix
291 - 310
outside
311 - 350
TMhelix
351 - 373
inside
374 - 385
TMhelix
386 - 408
outside
409 - 427
TMhelix
428 - 450
inside
451 - 468
TMhelix
469 - 491
outside
492 - 505
TMhelix
506 - 528
inside
529 - 580
Population Genetic Test Statistics
Pi
170.58769
Theta
162.895417
Tajima's D
-0.518604
CLR
176.288106
CSRT
0.23848807559622
Interpretation
Uncertain
