Gene
KWMTBOMO04906 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008066
Annotation
PREDICTED:_aminopeptidase_N-like_isoform_X1_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
PlasmaMembrane Reliability : 3.245
Sequence
CDS
ATGACACTATCATCGAGTCGTCACCAGTTTTTGGCGTTCGAATCGCAAGCTCCCGATGACATCCAATACAAAAGAAGAGGAGGGGTGTTCATTTCCCACTGTATCTGCGTAGCTTTCTTTATATTCGCCATTCTAACAGCTATCATTGTAGGAATAATAGCCCATTTCGTTACCTTTTATACGATATCAAATAAATCTACGGAATATTGGAATGAAGCCGAACCATTTGGGGTATCAGATGATAAACCATCACCAGATCTCAGACTACCCACATCGGTCACACCCAGCTTTTATCGTCTCAAAATTAAGGCAGATTTAGACAAAGCAAACTTCAGTGGAGATGTGCACATCACCTTAAAAGCGAATCGCAGAGTCAAAGAGATAATATTGCATTCCAAAGCACTGGTCATAAATAGTTCAAAGCTCACTGAACAAATATATGAAAGAGTTGAAACAATACACACCAAGGTGAAACGTGATGTACAAAATGAGAGTATTAGTGATACAACAACACCTGACAATAGTACCACAAATTTACCACCAACTACTGAAAGTCCGTCCCAAATTATCAATGAAACTACTGCTCCAATTATTACTGCTCCAATTATTACAACTCCAATTATTACTACTCCAATGATTACTACTCCAATTACTACTACAGCAAACCCAATATCAGTCGATACTCAAATAACACATAGCAGTGTACGCAGTGTAGAAATAATAGGCACATCTGTTGGTACTGGTGACCGATTGCTACTGACATTAGGTTCGGCTCTTACTCCTGGTGTGGATTACACTCTACAGCTATCATTCAGTGGAAATATAACAAATACACTAACTGGATTCTATAAGAGCACATATGTTGATAGCAACAATGAAAACAGATATTTAGGAGTAACTCAGTTTGAACCAACCTCAGCACGTTCAGTGTTCCCCTGCTTCGATGAGCCAGCATTCAAAGCCAAATTCGAAATAAGTATAGCACATCCTCAGAATTTAACTGTTTTATCCAACATGAAGGTAGCAACACAGGAGCCTATCACCGAAACACCGAAATGGCAATGGACGCACTTCGAACGTTCCGTGGATATGTCGACTTACCTTGTCGCTTTCGTGTTGTCCGACTTCACATCCCTAGAGACCAGTTACGTTAGCAAGGACAACGTAACGAAGCCAATACGGATATGGGCAAGGCCGGAACTGATAAGCAAAGCCAATTACGCCTTAAGAATCACACCGAAACTGCTCAATTATTACGAGGATGTTTTTGGAGTTCCGTACGTATTGGACAAACTAGACATGATCGCGATACCGGAATTCTCCAGCGGGGCCATGGAGAACTGGGGACTTATCACTTTCAGGGAAATGTCTTTGTTGTACGACGAAGCGGAAGGGATACCGCGCGACAAGCAAAACGTTGCGGTGTCCGTGGCCCACGAGCTCGCGCACCAGTGGTTCGGGAACCTCGTCACCATGCGCTGGTGGACGGACCTGTGGCTCAACGAGGGGTTCGCTTCATACATCGAGTACCTTGGAGTTGATCACATCGAACCGGAGTGGAATATGTTTGAGTCGTTCTCGAGAGATAAGATGGATCTCCTGCGATCGGACGCGCTGAAGAACACGTCCCCCGTCTCCAAGAAGGTGATGGACGCGTCGGAGATATCGCAGAAGTTTGACGAGATCTCGTACACGAAGGGCTCCAACTTGATCCGGATGTTGAACCACACGATATCCGAGCAACTCTTCCACAAGGGGCTGGTCATCTATCTGAACGACTGGAAATTCTCCAACGCGGAGGAAAACGACCTCTGGGCGGCCATGACTCGCGCCGTGAGCTCGGAGCGCGCGCCGGACGGCGAGTCCGTGGTGCGGCTCATGAACAGCTGGACGCGGCAGGCCGGCTACCCCGTCGTCACCGCCAACAGGAACTACGACACCGGCGCCGTCGAGATAGAGCAGAGATTGTTCACGAGCGCAAAGGACCCCTACCAATCGATGGTGGATCAGCTGTGGCACATTCCCATAAGCTACGTGAACGTGGACGCCCCGCTGGACGAGTGGAGCACCAAGCCCAAGACTTGGCTGAAGGACAGGATCAGCGTCTTCAACGTGCCCTACAACTCCACGCAGGCGCTGTACCTGAACGTCGATGCCATAGGCTACTACCGCGTGAACTACGACCAGAGGAACTGGCAGCTGCTGGCGGCGGCGCTGCGGGAGGGGCGGCTGCGGTCCGCCATCGCCGCGGCGCAGCTCGTCGACGACGCCTTCAACCTGGCGCGCGCCGCCCAGCTCGACTACGCGCACGCGCTGCAGCTGGCCGCCTGCGCCGCCGCCCGCCCCGGCCGCGTGCTGTGGGACCAGCTGCTCAACAACATGGCCGCGCTCAAGTACAACCTGATGACCACCGCCGGATACACTTACTTCCAAGACTTCATAAGGATACTGCTGAAGAACCAACTGGAGAGACTGAACTACGGCCTGGATAAGCCCAAGGACGACAACGAGGCTTTCCTTATCGAGAACCTGCTGATGTGGGAGTGCTACGCCGAGTCACCGCGCTGCCTGCGCTGGGCGCGTGCACAGTTCGACGCCTGGTACGCGCAGCACGACCACACCGCCATCCCGATCCCGAGCCACCTCCGCTCGCTGGTGCTGAACATGGCGCTGCGGCACGGCGGCCGCCAGGAGTTCGACTTCCTGTTCGAAGTGTTCCGCAACACCAGCGACCCCAGCCTCAAGGCGCTCATCATCAACAACCTGCCCAGCACCCGGGAGGAGAACCTCATCGTTCTGCTGCTGGAGAAGAGTCTGTCGGAGCTGCCGAAGCAGTACGCGGCGGCGGCGTGGGGCGTGGAGCCGGGGGCGGGGTCCCGGGTGGCGCAGGAGTACTTCTACCAGCACTTCGAGCGGATACACGCCCGGTTCGCCGACATGGACTCCTTCATCATGCCCACCGTGCTCAACGGGGCCTTCGGCTTCATCACCACCGACGACGAGCTGGCCAGGCTTAAAGCTTTCGCCGTGAAGCACAGGGAGCGGCTGCTGCCGGTGTCGCAGACGCTGCAGAAGCTGGTGGACACGGCGGCGCTGCGCATCCACTGGATCCGCAAGTACTCCGCCGGCATCGCGCGCTGGCTGCGGGACTACACGCAAGGTACACTTACAACGACCGAGGGCGGTGCAGTAAACACATCGACCACCGAAGCACCGGCGAACGCGACCATGGACATCAGCACGGCTCCACCGAGTACACCGAGTGCAGCTAGCAACACGACAACGTAA
Protein
MTLSSSRHQFLAFESQAPDDIQYKRRGGVFISHCICVAFFIFAILTAIIVGIIAHFVTFYTISNKSTEYWNEAEPFGVSDDKPSPDLRLPTSVTPSFYRLKIKADLDKANFSGDVHITLKANRRVKEIILHSKALVINSSKLTEQIYERVETIHTKVKRDVQNESISDTTTPDNSTTNLPPTTESPSQIINETTAPIITAPIITTPIITTPMITTPITTTANPISVDTQITHSSVRSVEIIGTSVGTGDRLLLTLGSALTPGVDYTLQLSFSGNITNTLTGFYKSTYVDSNNENRYLGVTQFEPTSARSVFPCFDEPAFKAKFEISIAHPQNLTVLSNMKVATQEPITETPKWQWTHFERSVDMSTYLVAFVLSDFTSLETSYVSKDNVTKPIRIWARPELISKANYALRITPKLLNYYEDVFGVPYVLDKLDMIAIPEFSSGAMENWGLITFREMSLLYDEAEGIPRDKQNVAVSVAHELAHQWFGNLVTMRWWTDLWLNEGFASYIEYLGVDHIEPEWNMFESFSRDKMDLLRSDALKNTSPVSKKVMDASEISQKFDEISYTKGSNLIRMLNHTISEQLFHKGLVIYLNDWKFSNAEENDLWAAMTRAVSSERAPDGESVVRLMNSWTRQAGYPVVTANRNYDTGAVEIEQRLFTSAKDPYQSMVDQLWHIPISYVNVDAPLDEWSTKPKTWLKDRISVFNVPYNSTQALYLNVDAIGYYRVNYDQRNWQLLAAALREGRLRSAIAAAQLVDDAFNLARAAQLDYAHALQLAACAAARPGRVLWDQLLNNMAALKYNLMTTAGYTYFQDFIRILLKNQLERLNYGLDKPKDDNEAFLIENLLMWECYAESPRCLRWARAQFDAWYAQHDHTAIPIPSHLRSLVLNMALRHGGRQEFDFLFEVFRNTSDPSLKALIINNLPSTREENLIVLLLEKSLSELPKQYAAAAWGVEPGAGSRVAQEYFYQHFERIHARFADMDSFIMPTVLNGAFGFITTDDELARLKAFAVKHRERLLPVSQTLQKLVDTAALRIHWIRKYSAGIARWLRDYTQGTLTTTEGGAVNTSTTEAPANATMDISTAPPSTPSAASNTTT
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Aminopeptidase
Uniprot
H9JEW9
A0A2H1W3N1
A0A2A4JG16
A0A194QR08
K7INE5
E2BB79
+ More
A0A2J7RAG3 A0A026W0P7 A0A195F0B1 A0A3L8E4L7 E9JC17 A0A232EWK0 E2ALT1 A0A2A3EJS2 A0A0L7QWL2 A0A067QSR5 A0A087ZUG8 A0A1E1XGK6 A0A195DN37 A0A158P3F3 A0A0M9A6N6 A0A146LYB5 A0A0P4VSA7 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 A0A0A1X3N6 E0VS71 A0A224Y1G3 A0A131XKB4 A0A131YLU7 A0A2R5LKG9 A0A034VBT7 A0A2L1IQ82 A0A310SPN7 A0A1B6GWM4 A0A0K8V993 A0A1I9WL50 W8BKE0 A0A0K8U897 A0A0P5EXD4 A0A0P5G2B7 A0A1B6CLC6 L7M3M3 A0A0P5JCR4 A0A0P5GQF7 A0A0P5KG78 A0A0P5PGC8 A0A0P6HT39 A0A1B6E6H9 A0A2H8TGE4 F4W4E5 A0A0N7ZUP7 A0A1B6M3I9 A0A2S2NGP8 T1I7S8 A0A0V0G738 A0A1Q3FSF6 A0A0P5I1L7 A0A3R7M3S3 X1WUZ5 A0A2R7VTS0 A0A1I8N5N4 A0A0P5GN39 A0A0P5H598 A0A182IV65 A0A0P5GP62 A0A0P5HT18 A0A0N8B1M1 A0A0L0BPV0 A0A084WR42 D6WCL5 A0A1B6D6B8 E9HM94 Q9VAP0 Q7KRW4 B3P5K7 A0A182T1W9 A0A2H1V5Y2 T1P8B3 A0A182GA80 B4HZ66 A0A2B4RUL2
A0A2J7RAG3 A0A026W0P7 A0A195F0B1 A0A3L8E4L7 E9JC17 A0A232EWK0 E2ALT1 A0A2A3EJS2 A0A0L7QWL2 A0A067QSR5 A0A087ZUG8 A0A1E1XGK6 A0A195DN37 A0A158P3F3 A0A0M9A6N6 A0A146LYB5 A0A0P4VSA7 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 A0A0A1X3N6 E0VS71 A0A224Y1G3 A0A131XKB4 A0A131YLU7 A0A2R5LKG9 A0A034VBT7 A0A2L1IQ82 A0A310SPN7 A0A1B6GWM4 A0A0K8V993 A0A1I9WL50 W8BKE0 A0A0K8U897 A0A0P5EXD4 A0A0P5G2B7 A0A1B6CLC6 L7M3M3 A0A0P5JCR4 A0A0P5GQF7 A0A0P5KG78 A0A0P5PGC8 A0A0P6HT39 A0A1B6E6H9 A0A2H8TGE4 F4W4E5 A0A0N7ZUP7 A0A1B6M3I9 A0A2S2NGP8 T1I7S8 A0A0V0G738 A0A1Q3FSF6 A0A0P5I1L7 A0A3R7M3S3 X1WUZ5 A0A2R7VTS0 A0A1I8N5N4 A0A0P5GN39 A0A0P5H598 A0A182IV65 A0A0P5GP62 A0A0P5HT18 A0A0N8B1M1 A0A0L0BPV0 A0A084WR42 D6WCL5 A0A1B6D6B8 E9HM94 Q9VAP0 Q7KRW4 B3P5K7 A0A182T1W9 A0A2H1V5Y2 T1P8B3 A0A182GA80 B4HZ66 A0A2B4RUL2
EC Number
3.4.11.-
Pubmed
19121390
26354079
20075255
20798317
24508170
30249741
+ More
21282665 28648823 24845553 28503490 21347285 26823975 27129103 26334808 25830018 20566863 28797301 28049606 26830274 25348373 29415259 27538518 24495485 25576852 21719571 25315136 26108605 24438588 18362917 19820115 21292972 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 26483478
21282665 28648823 24845553 28503490 21347285 26823975 27129103 26334808 25830018 20566863 28797301 28049606 26830274 25348373 29415259 27538518 24495485 25576852 21719571 25315136 26108605 24438588 18362917 19820115 21292972 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 26483478
EMBL
BABH01023421
JQ061150
AFK85024.1
ODYU01006102
SOQ47648.1
NWSH01001607
+ More
PCG70716.1 KQ461198 KPJ05966.1 GL446946 EFN87052.1 NEVH01006570 PNF37828.1 KK107589 EZA48634.1 KQ981905 KYN33597.1 QOIP01000001 RLU27566.1 GL771642 EFZ09701.1 NNAY01001852 OXU22728.1 GL440629 EFN65601.1 KZ288223 PBC31977.1 KQ414710 KOC62990.1 KK853596 KDR06435.1 GFAC01000979 JAT98209.1 KQ980713 KYN14241.1 ADTU01007781 ADTU01007782 ADTU01007783 KQ435724 KOX78330.1 GDHC01006045 JAQ12584.1 GDKW01001137 JAI55458.1 GBGD01000333 JAC88556.1 IACF01002668 LAB68313.1 GFTR01008371 JAW08055.1 GBXI01008343 JAD05949.1 DS235745 EEB16227.1 GFPF01000270 MAA11416.1 GEFH01001574 JAP67007.1 GEDV01009147 JAP79410.1 GGLE01005886 MBY10012.1 GAKP01019737 JAC39215.1 MF741655 AVD96938.1 KQ760397 OAD60666.1 GECZ01002996 JAS66773.1 GDHF01016887 JAI35427.1 KU932234 APA33870.1 GAMC01009092 JAB97463.1 GDHF01029538 JAI22776.1 GDIQ01268733 GDIQ01203036 GDIQ01126628 JAJ82991.1 GDIQ01247333 GDIQ01191687 JAK04392.1 GEDC01023029 GEDC01011996 JAS14269.1 JAS25302.1 GACK01007350 JAA57684.1 GDIQ01227787 GDIQ01226786 GDIQ01217985 GDIQ01164712 JAK24939.1 GDIQ01244546 JAK07179.1 GDIQ01241990 GDIQ01210238 GDIQ01190205 GDIQ01087933 GDIQ01086190 GDIQ01070141 JAK41487.1 GDIQ01150306 JAL01420.1 GDIQ01014599 JAN80138.1 GEDC01003748 JAS33550.1 GFXV01001305 MBW13110.1 GL887532 EGI70840.1 GDIP01210761 JAJ12641.1 GEBQ01009519 JAT30458.1 GGMR01003721 MBY16340.1 ACPB03005577 GECL01002882 JAP03242.1 GFDL01004639 JAV30406.1 GDIQ01225642 JAK26083.1 QCYY01002214 ROT72064.1 ABLF02030670 ABLF02030672 KK854013 PTY09335.1 GDIQ01238833 JAK12892.1 GDIQ01238832 JAK12893.1 GDIQ01238831 JAK12894.1 GDIQ01223140 JAK28585.1 GDIQ01221691 JAK30034.1 JRES01001565 KNC22036.1 ATLV01025918 KE525402 KFB52686.1 KQ971317 EEZ99313.2 GEDC01016064 JAS21234.1 GL732685 EFX67104.1 AE014297 AAF56863.2 AAN14161.2 CH954182 EDV53257.1 ODYU01000825 SOQ36186.1 KA644849 AFP59478.1 JXUM01050786 JXUM01050787 JXUM01050788 JXUM01050789 KQ561662 KXJ77885.1 CH480819 EDW53323.1 LSMT01000314 PFX20499.1
PCG70716.1 KQ461198 KPJ05966.1 GL446946 EFN87052.1 NEVH01006570 PNF37828.1 KK107589 EZA48634.1 KQ981905 KYN33597.1 QOIP01000001 RLU27566.1 GL771642 EFZ09701.1 NNAY01001852 OXU22728.1 GL440629 EFN65601.1 KZ288223 PBC31977.1 KQ414710 KOC62990.1 KK853596 KDR06435.1 GFAC01000979 JAT98209.1 KQ980713 KYN14241.1 ADTU01007781 ADTU01007782 ADTU01007783 KQ435724 KOX78330.1 GDHC01006045 JAQ12584.1 GDKW01001137 JAI55458.1 GBGD01000333 JAC88556.1 IACF01002668 LAB68313.1 GFTR01008371 JAW08055.1 GBXI01008343 JAD05949.1 DS235745 EEB16227.1 GFPF01000270 MAA11416.1 GEFH01001574 JAP67007.1 GEDV01009147 JAP79410.1 GGLE01005886 MBY10012.1 GAKP01019737 JAC39215.1 MF741655 AVD96938.1 KQ760397 OAD60666.1 GECZ01002996 JAS66773.1 GDHF01016887 JAI35427.1 KU932234 APA33870.1 GAMC01009092 JAB97463.1 GDHF01029538 JAI22776.1 GDIQ01268733 GDIQ01203036 GDIQ01126628 JAJ82991.1 GDIQ01247333 GDIQ01191687 JAK04392.1 GEDC01023029 GEDC01011996 JAS14269.1 JAS25302.1 GACK01007350 JAA57684.1 GDIQ01227787 GDIQ01226786 GDIQ01217985 GDIQ01164712 JAK24939.1 GDIQ01244546 JAK07179.1 GDIQ01241990 GDIQ01210238 GDIQ01190205 GDIQ01087933 GDIQ01086190 GDIQ01070141 JAK41487.1 GDIQ01150306 JAL01420.1 GDIQ01014599 JAN80138.1 GEDC01003748 JAS33550.1 GFXV01001305 MBW13110.1 GL887532 EGI70840.1 GDIP01210761 JAJ12641.1 GEBQ01009519 JAT30458.1 GGMR01003721 MBY16340.1 ACPB03005577 GECL01002882 JAP03242.1 GFDL01004639 JAV30406.1 GDIQ01225642 JAK26083.1 QCYY01002214 ROT72064.1 ABLF02030670 ABLF02030672 KK854013 PTY09335.1 GDIQ01238833 JAK12892.1 GDIQ01238832 JAK12893.1 GDIQ01238831 JAK12894.1 GDIQ01223140 JAK28585.1 GDIQ01221691 JAK30034.1 JRES01001565 KNC22036.1 ATLV01025918 KE525402 KFB52686.1 KQ971317 EEZ99313.2 GEDC01016064 JAS21234.1 GL732685 EFX67104.1 AE014297 AAF56863.2 AAN14161.2 CH954182 EDV53257.1 ODYU01000825 SOQ36186.1 KA644849 AFP59478.1 JXUM01050786 JXUM01050787 JXUM01050788 JXUM01050789 KQ561662 KXJ77885.1 CH480819 EDW53323.1 LSMT01000314 PFX20499.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000002358
UP000008237
UP000235965
+ More
UP000053097 UP000078541 UP000279307 UP000215335 UP000000311 UP000242457 UP000053825 UP000027135 UP000005203 UP000078492 UP000005205 UP000053105 UP000009046 UP000007755 UP000015103 UP000283509 UP000007819 UP000095301 UP000075880 UP000037069 UP000030765 UP000007266 UP000000305 UP000000803 UP000008711 UP000075901 UP000069940 UP000249989 UP000001292 UP000225706
UP000053097 UP000078541 UP000279307 UP000215335 UP000000311 UP000242457 UP000053825 UP000027135 UP000005203 UP000078492 UP000005205 UP000053105 UP000009046 UP000007755 UP000015103 UP000283509 UP000007819 UP000095301 UP000075880 UP000037069 UP000030765 UP000007266 UP000000305 UP000000803 UP000008711 UP000075901 UP000069940 UP000249989 UP000001292 UP000225706
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEW9
A0A2H1W3N1
A0A2A4JG16
A0A194QR08
K7INE5
E2BB79
+ More
A0A2J7RAG3 A0A026W0P7 A0A195F0B1 A0A3L8E4L7 E9JC17 A0A232EWK0 E2ALT1 A0A2A3EJS2 A0A0L7QWL2 A0A067QSR5 A0A087ZUG8 A0A1E1XGK6 A0A195DN37 A0A158P3F3 A0A0M9A6N6 A0A146LYB5 A0A0P4VSA7 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 A0A0A1X3N6 E0VS71 A0A224Y1G3 A0A131XKB4 A0A131YLU7 A0A2R5LKG9 A0A034VBT7 A0A2L1IQ82 A0A310SPN7 A0A1B6GWM4 A0A0K8V993 A0A1I9WL50 W8BKE0 A0A0K8U897 A0A0P5EXD4 A0A0P5G2B7 A0A1B6CLC6 L7M3M3 A0A0P5JCR4 A0A0P5GQF7 A0A0P5KG78 A0A0P5PGC8 A0A0P6HT39 A0A1B6E6H9 A0A2H8TGE4 F4W4E5 A0A0N7ZUP7 A0A1B6M3I9 A0A2S2NGP8 T1I7S8 A0A0V0G738 A0A1Q3FSF6 A0A0P5I1L7 A0A3R7M3S3 X1WUZ5 A0A2R7VTS0 A0A1I8N5N4 A0A0P5GN39 A0A0P5H598 A0A182IV65 A0A0P5GP62 A0A0P5HT18 A0A0N8B1M1 A0A0L0BPV0 A0A084WR42 D6WCL5 A0A1B6D6B8 E9HM94 Q9VAP0 Q7KRW4 B3P5K7 A0A182T1W9 A0A2H1V5Y2 T1P8B3 A0A182GA80 B4HZ66 A0A2B4RUL2
A0A2J7RAG3 A0A026W0P7 A0A195F0B1 A0A3L8E4L7 E9JC17 A0A232EWK0 E2ALT1 A0A2A3EJS2 A0A0L7QWL2 A0A067QSR5 A0A087ZUG8 A0A1E1XGK6 A0A195DN37 A0A158P3F3 A0A0M9A6N6 A0A146LYB5 A0A0P4VSA7 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 A0A0A1X3N6 E0VS71 A0A224Y1G3 A0A131XKB4 A0A131YLU7 A0A2R5LKG9 A0A034VBT7 A0A2L1IQ82 A0A310SPN7 A0A1B6GWM4 A0A0K8V993 A0A1I9WL50 W8BKE0 A0A0K8U897 A0A0P5EXD4 A0A0P5G2B7 A0A1B6CLC6 L7M3M3 A0A0P5JCR4 A0A0P5GQF7 A0A0P5KG78 A0A0P5PGC8 A0A0P6HT39 A0A1B6E6H9 A0A2H8TGE4 F4W4E5 A0A0N7ZUP7 A0A1B6M3I9 A0A2S2NGP8 T1I7S8 A0A0V0G738 A0A1Q3FSF6 A0A0P5I1L7 A0A3R7M3S3 X1WUZ5 A0A2R7VTS0 A0A1I8N5N4 A0A0P5GN39 A0A0P5H598 A0A182IV65 A0A0P5GP62 A0A0P5HT18 A0A0N8B1M1 A0A0L0BPV0 A0A084WR42 D6WCL5 A0A1B6D6B8 E9HM94 Q9VAP0 Q7KRW4 B3P5K7 A0A182T1W9 A0A2H1V5Y2 T1P8B3 A0A182GA80 B4HZ66 A0A2B4RUL2
PDB
5Z65
E-value=1.49252e-111,
Score=1034
Ontologies
PATHWAY
GO
PANTHER
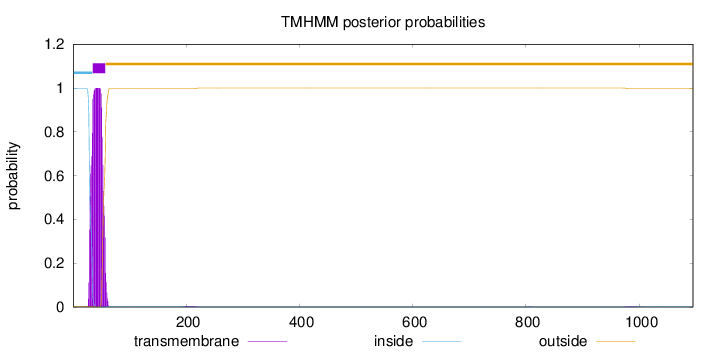
Topology
Length:
1095
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.1893999999999
Exp number, first 60 AAs:
24.00393
Total prob of N-in:
0.99711
POSSIBLE N-term signal
sequence
inside
1 - 34
TMhelix
35 - 57
outside
58 - 1095
Population Genetic Test Statistics
Pi
229.347745
Theta
165.340657
Tajima's D
1.195943
CLR
0.412041
CSRT
0.705164741762912
Interpretation
Uncertain
