Gene
KWMTBOMO04904
Pre Gene Modal
BGIBMGA008015
Annotation
aminopeptidase_N-like_precursor_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
Cytoplasmic Reliability : 3.316
Sequence
CDS
ATGAATATAGAAGTGATCCTGCTTGCTGCTCTGGGAGCCTGCGTGGCGTATTCCCTCCCACCGGAAGAGGTATCGGCTAGTCACATTATCAAAGCCAGAAGCATCGACCTCAATAACGCATATGGGCTCGCTATGGAAACCCGTTTGGAGAAGATCGTGGAGCCCACTGGATACAAGCTGGACTTAGAGCCGTTCCTCGACGACGGAGTCTACAGAGGAACCGTGAAAATCCAACTGAAGTGGCTGCAAGAGTCCGATGAGCTCAGCTTGCATTGTGACCATGAGCTTGGCATTAGCTTTTGGGACGTACAAGCGTATCCAGCGTCTGACGCGGAACATCCAGTAGAACGTGTCGTTGTGAAGGAGCTTCGTATGGACGTCAAAAAGCCGATATTGACTTTGTATTTCGAAAAGCCGATCCCCAAGGGAACCGAAGGACATATAGAGTTGACGTATCGAGGCAATATTCACATGGGTGTCACCGAGGGATTCTTCAAATCTACGTACACGACAGATCAGGGCGAAGAGATTATGGTAGCAGCTACACAGCTTCGTCCAAACAACGCGCGGCGTATGTTCCCCTGCTTCGACGAGCCGGGATACAAAACCCCGTTCGAAATCAGCGTAGCACGACCCCGGAACATGGTGGCCCTCAGCAATACTCCAATCGCGAGGACTGAGAATATAACCGGAGAACCGAATGCAGTGACGGATTACTTCGAGGTAACGCCTCCGATGTCCACGTTTACTGTTGGTCTAGTCATCGCCGATTTAAAGCAATTGGGGAACACTGTGCATTACAAAGATGACAATGGAAACGATATCGAACTGCGCGTGTGGGGACGAAAAGAATACCTTCAAGCGTTGGACGGTGTCGGTGAAAAATTCCTCAAAGTGTTCAGTGAGGTAGCGAACATGTGGCAAATTCCTTTACCGCTAAAGAAATTGGATATGGTCGCATTACCAAACTACCAGGGCGTCAGGCCGGCTGATAACTGGGGCCTGATAGTGTTCAAAGAGAGCGAGTTGAGCAACTGTAAAACTGTCCTTCTAGTCAATGAGGTACTGTACCAGTGGTTGGGAGTGTACGTGACTCCTGCTTGGTGGTCCGACGCTCATATAAACAAAGCCTTAGTCAATTCTTTGGCTGTTGAGATTTATTTTAAGATCAACAATGGCTCGGATATGGACGGAGACCGGCCCATCGGTGGCCTTTACTCTATTTACTATGAGTACAGCAAACGTTATCCGCATTCAAGAATAACTGGTATAAAGCAAGACGCTATTGCGACCAAAATAACATTGCTGTTCAGAACTTTGAACTATACACTCGGTGGAAATACACTGAAGAAAGGCTTAAGGACTTTCTTGCTTGAACATAAATACAAGACCTTTGTTGGTGACGATATATGGAATGCGCTAACTAAATCTGCTATTGACGACGGTAAAATCAGCAAAGACATAGATCTGAAAATTGTGGCCAAGAGCTGGATCGAGAAAGATCGATTGCCATTTCTGACTGTTGAAAGGAAGAATGATGGGAAAACTGTTGTGCTTACTCAGAAAGTATTCCTACGTGATCGTCCACACGACCTTCCAGACGCGGACAGGTTGACGTGGTGGCTCCCGGTTGTGATCGTAAGAGAGGATAATCTGGACTTCAGTAAAACTACCCCATTAGTTTGGATGAAGAACGTAAAGGAATTGACTATCAATGATATGCCAACCAAGGACAAGTTCATTATTATCAACCCCGAAGAGATCGCGCCTTACCTTGTGAACTACGACAAGGACACTTGGTATCTCTTATCCAAGTTCCTGCAGAGTGAAAAGCGCACACAGATCCCAGCTATAACCCGAGCCAAACTTTTACACGACGCATGGAATTTGGCTTATGCCGGTGAACTCTCCTTTGCCACTGCGCTGAATATGACTTTATTTTTGAAGACGGAAGAAGACCATTTGGTCTGGGAGGCTGTGTTTCCTATGCTGGATCATGTCGGAAGGCATATTTGTATGTGTATTCAAGGAAAATTCCAGGCATACGGTAGACATCTTTTGTTGCAAATCTATGAAAAACTAGGAAAAGAAGAGAAGGAAGGCGAAGAGAAATGGAAAACGAATCTGAGGTGCACTGTGAAAAGTTTTCTTTGCAGCAACGGGTACAAGCCTTGTGTCAAGGACGCCCAGGAACACTACGCTAAATGGATGCAAGCTAAGAATCCTGATGAAGGAAACCCAATTGCAAACCAGTACATTGGCCCGGTCTTCACGTACGGTACCAGAAAGGAATGGGAATTTGGCCTTCAGCGCGTCATCAACTTTCCTCCGTCGAGGAAGCAAAGTGAAAGAACTTACCTCCTGAAGACTTTAGCTGGCTGCCCCATTGAAGAGTACAAGATTAACAGGCTCCTGAACATAACGCTGCTGGAAGGAAACGGCAACTTCACTGATACGGATCTCTTCCTCATATTCAGTATGCTTACTGGTGGATCACAGGGGTACACGACCCTGTTCCATTTCTTGAACAACAATTGGGCAGTTTTAAAGGAGAAGTTCGCGAGCAAAACGAATCTATGGGACAGTCTGATTTCGGCAGCCACTGGACAATTCACGACCCAAGAGGGACTCGATCTAGTGTCCAACTTATACGTAGCTCACCAGGGCGAATTCGGGTCTGCCGAACACATCATAGAGACTTCCATGAAGAAGATACGCGAAGAAGCCAAATGGTCCGTCGAGAACATACCGGTCATAGAAAAATGGCTCGATGACTATGTAGCTAGATACGGAGTCAACGACGAATTGTTGGTTCAAAACAAATGA
Protein
MNIEVILLAALGACVAYSLPPEEVSASHIIKARSIDLNNAYGLAMETRLEKIVEPTGYKLDLEPFLDDGVYRGTVKIQLKWLQESDELSLHCDHELGISFWDVQAYPASDAEHPVERVVVKELRMDVKKPILTLYFEKPIPKGTEGHIELTYRGNIHMGVTEGFFKSTYTTDQGEEIMVAATQLRPNNARRMFPCFDEPGYKTPFEISVARPRNMVALSNTPIARTENITGEPNAVTDYFEVTPPMSTFTVGLVIADLKQLGNTVHYKDDNGNDIELRVWGRKEYLQALDGVGEKFLKVFSEVANMWQIPLPLKKLDMVALPNYQGVRPADNWGLIVFKESELSNCKTVLLVNEVLYQWLGVYVTPAWWSDAHINKALVNSLAVEIYFKINNGSDMDGDRPIGGLYSIYYEYSKRYPHSRITGIKQDAIATKITLLFRTLNYTLGGNTLKKGLRTFLLEHKYKTFVGDDIWNALTKSAIDDGKISKDIDLKIVAKSWIEKDRLPFLTVERKNDGKTVVLTQKVFLRDRPHDLPDADRLTWWLPVVIVREDNLDFSKTTPLVWMKNVKELTINDMPTKDKFIIINPEEIAPYLVNYDKDTWYLLSKFLQSEKRTQIPAITRAKLLHDAWNLAYAGELSFATALNMTLFLKTEEDHLVWEAVFPMLDHVGRHICMCIQGKFQAYGRHLLLQIYEKLGKEEKEGEEKWKTNLRCTVKSFLCSNGYKPCVKDAQEHYAKWMQAKNPDEGNPIANQYIGPVFTYGTRKEWEFGLQRVINFPPSRKQSERTYLLKTLAGCPIEEYKINRLLNITLLEGNGNFTDTDLFLIFSMLTGGSQGYTTLFHFLNNNWAVLKEKFASKTNLWDSLISAATGQFTTQEGLDLVSNLYVAHQGEFGSAEHIIETSMKKIREEAKWSVENIPVIEKWLDDYVARYGVNDELLVQNK
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Uniprot
H9JER8
A0A2A4JF85
A0A2W1BHJ6
A0A194QIT7
A0A194QKA5
A0A3S2L2I0
+ More
A0A1J1J0P9 D6WCP6 A0A1Y1MA14 A0A182YQT4 A0A182MTI3 A0A336MBD0 A0A182PUA0 Q7PRZ0 A0A182HIH8 A0A182W435 A0A084WPH3 A0A182RVV1 A0A182VJH1 U5EY25 A0A182NAG4 A0A182QEL8 A0A182IM68 A0A212F119 A0A1Q3FQV7 B0WWU9 A0A182G9P3 W5JK35 A0A1S4FVA8 Q16N34 A0A0T6B5L3 B4NIY5 A0A1W4WRQ9 A0A182WY93 A0A1W4X1D8 A0A2M4AFZ3 A0A1I8NZK0 A0A1A9WGS8 A0A2M4AHH4 A0A0Q9X861 A0A1A9VNN8 A0A0Q9W942 A0A1B0CUZ1 A0A1B0GDF9 A0A1A9ZHW6 A0A1A9YBD6 A0A1B0AZS0 A0A182KDE0 A0A3B0KHT9 A0A154PMP7 A0A087ZWN2 A0A2A3E6Y7 V9I6X5 A0A310SID4 Q7PLV6 A0A0N0BKZ8 F4WPN4 E2C7E9 T1PI34 A0A158P1C9 K7IND9 I5AP64 A0A195ESI7 A0A0C9QHW8 A0A026X190 A0A151XHE3 A0A195DUN3 A0A151IEB3 A0A232EMF5 A0A067QK30 E2AGD8 Q500Y3 A0A195BLM7 A0A0J9US13 A0A0L7RE38 A0A2J7RK94 A0A182SLF0 A0A0L0CG78 A0A2J7RK98 A0A1B6BZX2 A0A1B6CI33 A0A0A9XHA9 T1HEW7 A0A0A9Y597 A0A1B6KBG8 A0A2A3EJS2
A0A1J1J0P9 D6WCP6 A0A1Y1MA14 A0A182YQT4 A0A182MTI3 A0A336MBD0 A0A182PUA0 Q7PRZ0 A0A182HIH8 A0A182W435 A0A084WPH3 A0A182RVV1 A0A182VJH1 U5EY25 A0A182NAG4 A0A182QEL8 A0A182IM68 A0A212F119 A0A1Q3FQV7 B0WWU9 A0A182G9P3 W5JK35 A0A1S4FVA8 Q16N34 A0A0T6B5L3 B4NIY5 A0A1W4WRQ9 A0A182WY93 A0A1W4X1D8 A0A2M4AFZ3 A0A1I8NZK0 A0A1A9WGS8 A0A2M4AHH4 A0A0Q9X861 A0A1A9VNN8 A0A0Q9W942 A0A1B0CUZ1 A0A1B0GDF9 A0A1A9ZHW6 A0A1A9YBD6 A0A1B0AZS0 A0A182KDE0 A0A3B0KHT9 A0A154PMP7 A0A087ZWN2 A0A2A3E6Y7 V9I6X5 A0A310SID4 Q7PLV6 A0A0N0BKZ8 F4WPN4 E2C7E9 T1PI34 A0A158P1C9 K7IND9 I5AP64 A0A195ESI7 A0A0C9QHW8 A0A026X190 A0A151XHE3 A0A195DUN3 A0A151IEB3 A0A232EMF5 A0A067QK30 E2AGD8 Q500Y3 A0A195BLM7 A0A0J9US13 A0A0L7RE38 A0A2J7RK94 A0A182SLF0 A0A0L0CG78 A0A2J7RK98 A0A1B6BZX2 A0A1B6CI33 A0A0A9XHA9 T1HEW7 A0A0A9Y597 A0A1B6KBG8 A0A2A3EJS2
EC Number
3.4.11.-
Pubmed
19121390
28756777
26354079
18362917
19820115
28004739
+ More
25244985 12364791 14747013 17210077 24438588 22118469 26483478 20920257 23761445 17510324 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 20798317 25315136 21347285 20075255 15632085 24508170 30249741 28648823 24845553 22936249 26108605 25401762
25244985 12364791 14747013 17210077 24438588 22118469 26483478 20920257 23761445 17510324 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 20798317 25315136 21347285 20075255 15632085 24508170 30249741 28648823 24845553 22936249 26108605 25401762
EMBL
BABH01023418
JQ061156
AFK85030.1
NWSH01001607
PCG70717.1
KZ150199
+ More
PZC72296.1 KQ459185 KPJ03366.1 KQ461198 KPJ05967.1 RSAL01000211 RVE44187.1 CVRI01000066 CRL06029.1 KQ971317 EEZ99170.1 GEZM01036661 JAV82534.1 AXCM01000646 UFQT01000650 SSX26113.1 AAAB01008846 EAA06384.5 APCN01003972 ATLV01025077 KE525369 KFB52117.1 GANO01000587 JAB59284.1 AXCN02002022 AXCP01007276 AGBW02011001 OWR47404.1 GFDL01005096 JAV29949.1 DS232152 EDS36210.1 JXUM01049614 KQ561613 KXJ78002.1 ADMH02001274 ETN63260.1 CH477841 EAT35752.1 LJIG01009638 KRT82680.1 CH964272 EDW84887.2 GGFK01006394 MBW39715.1 GGFK01006909 MBW40230.1 CH933806 KRG00711.1 KRG00712.1 KRG00713.1 CH940657 KRF80934.1 KRF80935.1 AJWK01029849 CCAG010011873 JXJN01006448 OUUW01000007 SPP83268.1 KQ434984 KZC13129.1 KZ288354 PBC27264.1 JR035962 JR035963 AEY56828.1 AEY56829.1 KQ763442 OAD55092.1 BT120390 AE014296 ADD20764.1 EAA46083.2 KQ435691 KOX81164.1 GL888251 EGI63838.1 GL453369 EFN76136.1 KA648426 AFP63055.1 ADTU01006291 ADTU01006292 CM000070 EIM52749.2 KQ981989 KYN31136.1 GBYB01003139 JAG72906.1 KK107046 QOIP01000004 EZA61776.1 RLU23236.1 KQ982138 KYQ59630.1 KQ980390 KYN16264.1 KQ977877 KYM99029.1 NNAY01003376 OXU19521.1 KK853256 KDR09258.1 GL439310 EFN67471.1 BT022084 AAY33500.1 KQ976449 KYM85819.1 CM002912 KMZ01331.1 KQ414612 KOC69113.1 NEVH01002981 PNF41264.1 JRES01000426 KNC31388.1 PNF41265.1 GEDC01030773 JAS06525.1 GEDC01024221 GEDC01015585 JAS13077.1 JAS21713.1 GBHO01025429 GBHO01025426 GBRD01008812 JAG18175.1 JAG18178.1 JAG57009.1 ACPB03020693 GBHO01025428 GBHO01016255 JAG18176.1 JAG27349.1 GEBQ01031187 JAT08790.1 KZ288223 PBC31977.1
PZC72296.1 KQ459185 KPJ03366.1 KQ461198 KPJ05967.1 RSAL01000211 RVE44187.1 CVRI01000066 CRL06029.1 KQ971317 EEZ99170.1 GEZM01036661 JAV82534.1 AXCM01000646 UFQT01000650 SSX26113.1 AAAB01008846 EAA06384.5 APCN01003972 ATLV01025077 KE525369 KFB52117.1 GANO01000587 JAB59284.1 AXCN02002022 AXCP01007276 AGBW02011001 OWR47404.1 GFDL01005096 JAV29949.1 DS232152 EDS36210.1 JXUM01049614 KQ561613 KXJ78002.1 ADMH02001274 ETN63260.1 CH477841 EAT35752.1 LJIG01009638 KRT82680.1 CH964272 EDW84887.2 GGFK01006394 MBW39715.1 GGFK01006909 MBW40230.1 CH933806 KRG00711.1 KRG00712.1 KRG00713.1 CH940657 KRF80934.1 KRF80935.1 AJWK01029849 CCAG010011873 JXJN01006448 OUUW01000007 SPP83268.1 KQ434984 KZC13129.1 KZ288354 PBC27264.1 JR035962 JR035963 AEY56828.1 AEY56829.1 KQ763442 OAD55092.1 BT120390 AE014296 ADD20764.1 EAA46083.2 KQ435691 KOX81164.1 GL888251 EGI63838.1 GL453369 EFN76136.1 KA648426 AFP63055.1 ADTU01006291 ADTU01006292 CM000070 EIM52749.2 KQ981989 KYN31136.1 GBYB01003139 JAG72906.1 KK107046 QOIP01000004 EZA61776.1 RLU23236.1 KQ982138 KYQ59630.1 KQ980390 KYN16264.1 KQ977877 KYM99029.1 NNAY01003376 OXU19521.1 KK853256 KDR09258.1 GL439310 EFN67471.1 BT022084 AAY33500.1 KQ976449 KYM85819.1 CM002912 KMZ01331.1 KQ414612 KOC69113.1 NEVH01002981 PNF41264.1 JRES01000426 KNC31388.1 PNF41265.1 GEDC01030773 JAS06525.1 GEDC01024221 GEDC01015585 JAS13077.1 JAS21713.1 GBHO01025429 GBHO01025426 GBRD01008812 JAG18175.1 JAG18178.1 JAG57009.1 ACPB03020693 GBHO01025428 GBHO01016255 JAG18176.1 JAG27349.1 GEBQ01031187 JAT08790.1 KZ288223 PBC31977.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000183832
+ More
UP000007266 UP000076408 UP000075883 UP000075885 UP000007062 UP000075840 UP000075920 UP000030765 UP000075900 UP000075903 UP000075884 UP000075886 UP000075880 UP000007151 UP000002320 UP000069940 UP000249989 UP000000673 UP000008820 UP000007798 UP000192223 UP000076407 UP000095300 UP000091820 UP000009192 UP000078200 UP000008792 UP000092461 UP000092444 UP000092445 UP000092443 UP000092460 UP000075881 UP000268350 UP000076502 UP000005203 UP000242457 UP000000803 UP000053105 UP000007755 UP000008237 UP000095301 UP000005205 UP000002358 UP000001819 UP000078541 UP000053097 UP000279307 UP000075809 UP000078492 UP000078542 UP000215335 UP000027135 UP000000311 UP000078540 UP000053825 UP000235965 UP000075901 UP000037069 UP000015103
UP000007266 UP000076408 UP000075883 UP000075885 UP000007062 UP000075840 UP000075920 UP000030765 UP000075900 UP000075903 UP000075884 UP000075886 UP000075880 UP000007151 UP000002320 UP000069940 UP000249989 UP000000673 UP000008820 UP000007798 UP000192223 UP000076407 UP000095300 UP000091820 UP000009192 UP000078200 UP000008792 UP000092461 UP000092444 UP000092445 UP000092443 UP000092460 UP000075881 UP000268350 UP000076502 UP000005203 UP000242457 UP000000803 UP000053105 UP000007755 UP000008237 UP000095301 UP000005205 UP000002358 UP000001819 UP000078541 UP000053097 UP000279307 UP000075809 UP000078492 UP000078542 UP000215335 UP000027135 UP000000311 UP000078540 UP000053825 UP000235965 UP000075901 UP000037069 UP000015103
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JER8
A0A2A4JF85
A0A2W1BHJ6
A0A194QIT7
A0A194QKA5
A0A3S2L2I0
+ More
A0A1J1J0P9 D6WCP6 A0A1Y1MA14 A0A182YQT4 A0A182MTI3 A0A336MBD0 A0A182PUA0 Q7PRZ0 A0A182HIH8 A0A182W435 A0A084WPH3 A0A182RVV1 A0A182VJH1 U5EY25 A0A182NAG4 A0A182QEL8 A0A182IM68 A0A212F119 A0A1Q3FQV7 B0WWU9 A0A182G9P3 W5JK35 A0A1S4FVA8 Q16N34 A0A0T6B5L3 B4NIY5 A0A1W4WRQ9 A0A182WY93 A0A1W4X1D8 A0A2M4AFZ3 A0A1I8NZK0 A0A1A9WGS8 A0A2M4AHH4 A0A0Q9X861 A0A1A9VNN8 A0A0Q9W942 A0A1B0CUZ1 A0A1B0GDF9 A0A1A9ZHW6 A0A1A9YBD6 A0A1B0AZS0 A0A182KDE0 A0A3B0KHT9 A0A154PMP7 A0A087ZWN2 A0A2A3E6Y7 V9I6X5 A0A310SID4 Q7PLV6 A0A0N0BKZ8 F4WPN4 E2C7E9 T1PI34 A0A158P1C9 K7IND9 I5AP64 A0A195ESI7 A0A0C9QHW8 A0A026X190 A0A151XHE3 A0A195DUN3 A0A151IEB3 A0A232EMF5 A0A067QK30 E2AGD8 Q500Y3 A0A195BLM7 A0A0J9US13 A0A0L7RE38 A0A2J7RK94 A0A182SLF0 A0A0L0CG78 A0A2J7RK98 A0A1B6BZX2 A0A1B6CI33 A0A0A9XHA9 T1HEW7 A0A0A9Y597 A0A1B6KBG8 A0A2A3EJS2
A0A1J1J0P9 D6WCP6 A0A1Y1MA14 A0A182YQT4 A0A182MTI3 A0A336MBD0 A0A182PUA0 Q7PRZ0 A0A182HIH8 A0A182W435 A0A084WPH3 A0A182RVV1 A0A182VJH1 U5EY25 A0A182NAG4 A0A182QEL8 A0A182IM68 A0A212F119 A0A1Q3FQV7 B0WWU9 A0A182G9P3 W5JK35 A0A1S4FVA8 Q16N34 A0A0T6B5L3 B4NIY5 A0A1W4WRQ9 A0A182WY93 A0A1W4X1D8 A0A2M4AFZ3 A0A1I8NZK0 A0A1A9WGS8 A0A2M4AHH4 A0A0Q9X861 A0A1A9VNN8 A0A0Q9W942 A0A1B0CUZ1 A0A1B0GDF9 A0A1A9ZHW6 A0A1A9YBD6 A0A1B0AZS0 A0A182KDE0 A0A3B0KHT9 A0A154PMP7 A0A087ZWN2 A0A2A3E6Y7 V9I6X5 A0A310SID4 Q7PLV6 A0A0N0BKZ8 F4WPN4 E2C7E9 T1PI34 A0A158P1C9 K7IND9 I5AP64 A0A195ESI7 A0A0C9QHW8 A0A026X190 A0A151XHE3 A0A195DUN3 A0A151IEB3 A0A232EMF5 A0A067QK30 E2AGD8 Q500Y3 A0A195BLM7 A0A0J9US13 A0A0L7RE38 A0A2J7RK94 A0A182SLF0 A0A0L0CG78 A0A2J7RK98 A0A1B6BZX2 A0A1B6CI33 A0A0A9XHA9 T1HEW7 A0A0A9Y597 A0A1B6KBG8 A0A2A3EJS2
PDB
6Q4R
E-value=1.01883e-60,
Score=595
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.959300
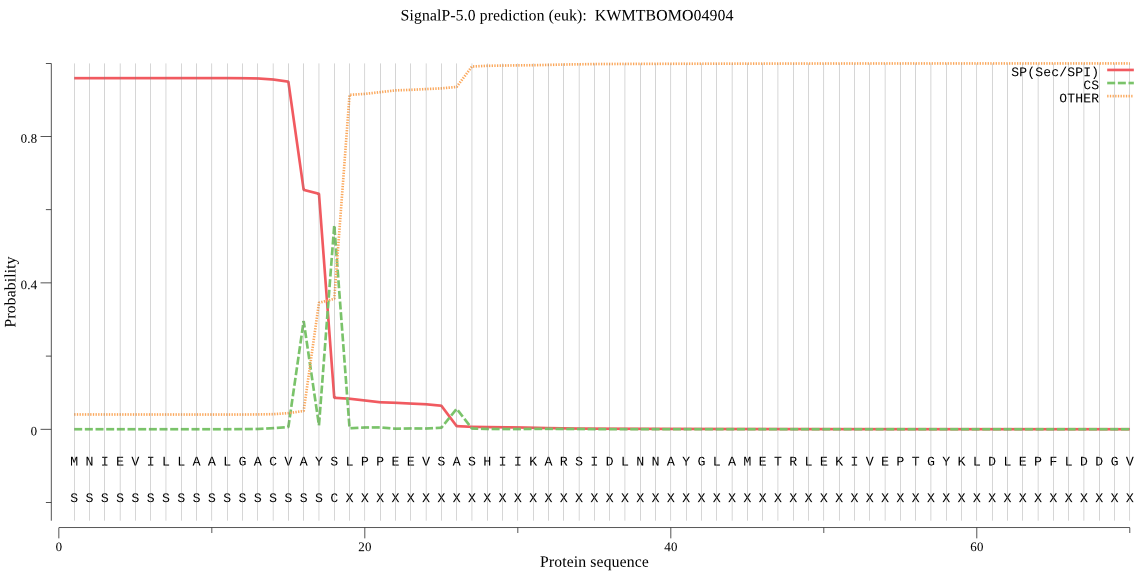
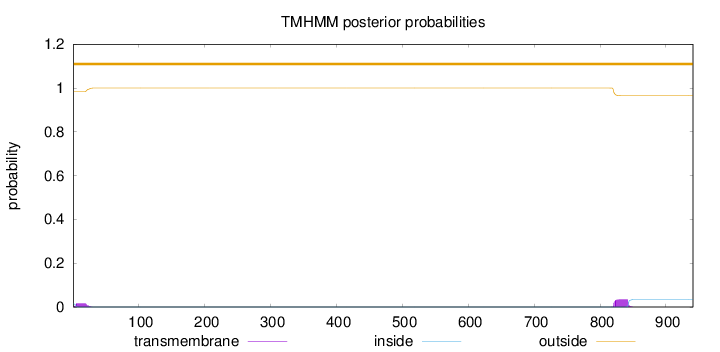
Length:
941
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.09683
Exp number, first 60 AAs:
0.31616
Total prob of N-in:
0.01670
outside
1 - 941
Population Genetic Test Statistics
Pi
217.828196
Theta
147.643271
Tajima's D
1.503727
CLR
0.344532
CSRT
0.787960601969901
Interpretation
Uncertain
