Gene
KWMTBOMO04903
Pre Gene Modal
BGIBMGA008016
Annotation
PREDICTED:_WAP_four-disulfide_core_domain_protein_2-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 4.037
Sequence
CDS
ATGACCCGTGAGTTCAAGAGTCCAGAAATGGTGCGTTGTTTACTTCTAATTATCGTGTCCTGCCTCACATCGTACTCGCTGTGCGGTTCGTGTCCTCCGACGTTGTCCGTGGACATTTGCGAGCCAATGTGTGGTCCCGGTCAGGAGTGTAACGGGACTCAGCTCTGCTGTCCCACGCACTGCGGTGGAGCCATGTGTGTTGATGCGATGACTCAGAGGCACTTTGTTCATTTAGTAAAGAAAGGTAATTGCCCCGAGTTTCCGCGCGGCCCGTGGATCTGCTCCCACACTTGCACCGGCGACTCGGACTGCCCTCGTGCCCTCAAGTGCTGTCACAACCGCTGCGGGGTCCTCACCTGTCAGAAGCCGGAAATAGATCCCGAACCATTTGTTGAACTTCCATAG
Protein
MTREFKSPEMVRCLLLIIVSCLTSYSLCGSCPPTLSVDICEPMCGPGQECNGTQLCCPTHCGGAMCVDAMTQRHFVHLVKKGNCPEFPRGPWICSHTCTGDSDCPRALKCCHNRCGVLTCQKPEIDPEPFVELP
Summary
Uniprot
H9JER9
A0A194QCV0
A0A194QK69
I4DJU3
A0A2W1BGG8
A0A2A4JFX1
+ More
A0A212EU94 A0A194QKA5 A0A3S2NNA5 A0A194QD28 S4NYM1 A0A2H1WUT2 A0A0L7KUT3 E2AH82 A0A067QSE3 A0A087ZR76 A0A0L7QQK8 A0A1B6F003 E9IH86 A0A026WK52 A0A023F117 A0A154PHF9 A0A1B6HND8 A0A195DMZ1 A0A348G5X9 E2BFY2 A0A195BJB1 A0A158NA97 A0A0V0G611 A0A0N7Z8I4 V9IL21 A0A195FRB7 T1HUX6 A0A151X7Q4 A0A2A3E4R2 A0A0J7KMP0 D6X3Z5 A0A2K8JSP6 F4WN59 A0A224Y1P5 A0A0T6B9Q1 A0A195CBH9 A0A2J7PDN1 A0A0C9QP19 A0A1B6DJH2 A0A1W4WH03 A0A0M9ACY7 A0A1Y1LHQ0 A0A1Y1ME15 A0A146L0X7 A0A3B3R0H1 A0A1Y1N025 A0A0N5DZH8 A0A2D1QU04 A0A2D1QU21 A0A2D1QTX3 A0A1S3MF07 A0A3B3ZGR4 H3BH40 A0A3P8ZQ78 A0A210QLV6 G3UUY2 A0A3B3ZGS3 A0A2D1QTZ2 A0A3M7S2V3 B5XAK3 B5XFI6 A0A2G8KFG3 A0A151M3K8 A0A2G8JL59 A0A3B3ZH74 V8N9T5 A0A3N0XW44 A0A3B3ZGN5
A0A212EU94 A0A194QKA5 A0A3S2NNA5 A0A194QD28 S4NYM1 A0A2H1WUT2 A0A0L7KUT3 E2AH82 A0A067QSE3 A0A087ZR76 A0A0L7QQK8 A0A1B6F003 E9IH86 A0A026WK52 A0A023F117 A0A154PHF9 A0A1B6HND8 A0A195DMZ1 A0A348G5X9 E2BFY2 A0A195BJB1 A0A158NA97 A0A0V0G611 A0A0N7Z8I4 V9IL21 A0A195FRB7 T1HUX6 A0A151X7Q4 A0A2A3E4R2 A0A0J7KMP0 D6X3Z5 A0A2K8JSP6 F4WN59 A0A224Y1P5 A0A0T6B9Q1 A0A195CBH9 A0A2J7PDN1 A0A0C9QP19 A0A1B6DJH2 A0A1W4WH03 A0A0M9ACY7 A0A1Y1LHQ0 A0A1Y1ME15 A0A146L0X7 A0A3B3R0H1 A0A1Y1N025 A0A0N5DZH8 A0A2D1QU04 A0A2D1QU21 A0A2D1QTX3 A0A1S3MF07 A0A3B3ZGR4 H3BH40 A0A3P8ZQ78 A0A210QLV6 G3UUY2 A0A3B3ZGS3 A0A2D1QTZ2 A0A3M7S2V3 B5XAK3 B5XFI6 A0A2G8KFG3 A0A151M3K8 A0A2G8JL59 A0A3B3ZH74 V8N9T5 A0A3N0XW44 A0A3B3ZGN5
Pubmed
EMBL
BABH01023416
BABH01023417
KQ459185
KPJ03368.1
KQ461198
KPJ05968.1
+ More
AK401561 BAM18183.1 KZ150199 PZC72297.1 NWSH01001607 PCG70719.1 AGBW02012440 OWR45058.1 KPJ05967.1 RSAL01000211 RVE44186.1 KPJ03367.1 GAIX01013765 JAA78795.1 ODYU01011213 SOQ56813.1 JTDY01005660 KOB66774.1 GL439483 EFN67199.1 KK853353 KDR08241.1 KQ414786 KOC60912.1 GECZ01026133 JAS43636.1 GL763190 EFZ20060.1 KK107167 QOIP01000010 EZA56400.1 RLU18039.1 GBBI01004001 JAC14711.1 KQ434896 KZC10758.1 GECU01031517 GECU01021139 JAS76189.1 JAS86567.1 KQ980734 KYN13869.1 FX985516 BBF97852.1 GL448096 EFN85384.1 KQ976455 KYM85186.1 ADTU01010081 GECL01002601 JAP03523.1 GDKW01003259 JAI53336.1 JR052634 AEY61745.1 KQ981305 KYN43008.1 ACPB03010126 KQ982446 KYQ56354.1 KZ288375 PBC26678.1 LBMM01005297 KMQ91638.1 KQ971379 EEZ97731.1 KY031319 ATU83070.1 GL888235 EGI64328.1 GFTR01002087 JAW14339.1 LJIG01009174 KRT83607.1 KQ978009 KYM98157.1 NEVH01026386 PNF14445.1 GBYB01002397 JAG72164.1 GEDC01011496 JAS25802.1 KQ435699 KOX80599.1 GEZM01059401 JAV71495.1 GEZM01036280 GEZM01036279 JAV82830.1 GDHC01017010 JAQ01619.1 GEZM01019660 JAV89586.1 MF371325 ATP16009.1 MF371326 ATP16010.1 MF371323 ATP16007.1 AFYH01003877 NEDP02002985 OWF49718.1 MF371324 ATP16008.1 REGN01002122 RNA30102.1 BT048072 ACI67873.1 BT049805 ACI69606.1 MRZV01000626 PIK46710.1 AKHW03006757 KYO19109.1 MRZV01001671 PIK36460.1 AZIM01006891 ETE58332.1 RJVU01060476 ROJ48034.1
AK401561 BAM18183.1 KZ150199 PZC72297.1 NWSH01001607 PCG70719.1 AGBW02012440 OWR45058.1 KPJ05967.1 RSAL01000211 RVE44186.1 KPJ03367.1 GAIX01013765 JAA78795.1 ODYU01011213 SOQ56813.1 JTDY01005660 KOB66774.1 GL439483 EFN67199.1 KK853353 KDR08241.1 KQ414786 KOC60912.1 GECZ01026133 JAS43636.1 GL763190 EFZ20060.1 KK107167 QOIP01000010 EZA56400.1 RLU18039.1 GBBI01004001 JAC14711.1 KQ434896 KZC10758.1 GECU01031517 GECU01021139 JAS76189.1 JAS86567.1 KQ980734 KYN13869.1 FX985516 BBF97852.1 GL448096 EFN85384.1 KQ976455 KYM85186.1 ADTU01010081 GECL01002601 JAP03523.1 GDKW01003259 JAI53336.1 JR052634 AEY61745.1 KQ981305 KYN43008.1 ACPB03010126 KQ982446 KYQ56354.1 KZ288375 PBC26678.1 LBMM01005297 KMQ91638.1 KQ971379 EEZ97731.1 KY031319 ATU83070.1 GL888235 EGI64328.1 GFTR01002087 JAW14339.1 LJIG01009174 KRT83607.1 KQ978009 KYM98157.1 NEVH01026386 PNF14445.1 GBYB01002397 JAG72164.1 GEDC01011496 JAS25802.1 KQ435699 KOX80599.1 GEZM01059401 JAV71495.1 GEZM01036280 GEZM01036279 JAV82830.1 GDHC01017010 JAQ01619.1 GEZM01019660 JAV89586.1 MF371325 ATP16009.1 MF371326 ATP16010.1 MF371323 ATP16007.1 AFYH01003877 NEDP02002985 OWF49718.1 MF371324 ATP16008.1 REGN01002122 RNA30102.1 BT048072 ACI67873.1 BT049805 ACI69606.1 MRZV01000626 PIK46710.1 AKHW03006757 KYO19109.1 MRZV01001671 PIK36460.1 AZIM01006891 ETE58332.1 RJVU01060476 ROJ48034.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000283053
+ More
UP000037510 UP000000311 UP000027135 UP000005203 UP000053825 UP000053097 UP000279307 UP000076502 UP000078492 UP000008237 UP000078540 UP000005205 UP000078541 UP000015103 UP000075809 UP000242457 UP000036403 UP000007266 UP000007755 UP000078542 UP000235965 UP000192223 UP000053105 UP000261540 UP000046395 UP000087266 UP000261520 UP000008672 UP000265140 UP000242188 UP000001645 UP000276133 UP000230750 UP000050525
UP000037510 UP000000311 UP000027135 UP000005203 UP000053825 UP000053097 UP000279307 UP000076502 UP000078492 UP000008237 UP000078540 UP000005205 UP000078541 UP000015103 UP000075809 UP000242457 UP000036403 UP000007266 UP000007755 UP000078542 UP000235965 UP000192223 UP000053105 UP000261540 UP000046395 UP000087266 UP000261520 UP000008672 UP000265140 UP000242188 UP000001645 UP000276133 UP000230750 UP000050525
PRIDE
Pfam
Interpro
IPR036645
Elafin-like_sf
+ More
IPR008197 WAP_dom
IPR001930 Peptidase_M1
IPR042097 Aminopeptidase_N-like_N
IPR024571 ERAP1-like_C_dom
IPR014782 Peptidase_M1_dom
IPR008037 Pacifastin_dom
IPR006150 Cys_repeat_1
IPR002223 Kunitz_BPTI
IPR000716 Thyroglobulin_1
IPR036857 Thyroglobulin_1_sf
IPR036880 Kunitz_BPTI_sf
IPR029716 Wfdc18
IPR020901 Prtase_inh_Kunz-CS
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013106 Ig_V-set
IPR013098 Ig_I-set
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR011061 Hirudin/antistatin
IPR004094 Antistasin-like
IPR008197 WAP_dom
IPR001930 Peptidase_M1
IPR042097 Aminopeptidase_N-like_N
IPR024571 ERAP1-like_C_dom
IPR014782 Peptidase_M1_dom
IPR008037 Pacifastin_dom
IPR006150 Cys_repeat_1
IPR002223 Kunitz_BPTI
IPR000716 Thyroglobulin_1
IPR036857 Thyroglobulin_1_sf
IPR036880 Kunitz_BPTI_sf
IPR029716 Wfdc18
IPR020901 Prtase_inh_Kunz-CS
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013106 Ig_V-set
IPR013098 Ig_I-set
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR011061 Hirudin/antistatin
IPR004094 Antistasin-like
SUPFAM
CDD
ProteinModelPortal
H9JER9
A0A194QCV0
A0A194QK69
I4DJU3
A0A2W1BGG8
A0A2A4JFX1
+ More
A0A212EU94 A0A194QKA5 A0A3S2NNA5 A0A194QD28 S4NYM1 A0A2H1WUT2 A0A0L7KUT3 E2AH82 A0A067QSE3 A0A087ZR76 A0A0L7QQK8 A0A1B6F003 E9IH86 A0A026WK52 A0A023F117 A0A154PHF9 A0A1B6HND8 A0A195DMZ1 A0A348G5X9 E2BFY2 A0A195BJB1 A0A158NA97 A0A0V0G611 A0A0N7Z8I4 V9IL21 A0A195FRB7 T1HUX6 A0A151X7Q4 A0A2A3E4R2 A0A0J7KMP0 D6X3Z5 A0A2K8JSP6 F4WN59 A0A224Y1P5 A0A0T6B9Q1 A0A195CBH9 A0A2J7PDN1 A0A0C9QP19 A0A1B6DJH2 A0A1W4WH03 A0A0M9ACY7 A0A1Y1LHQ0 A0A1Y1ME15 A0A146L0X7 A0A3B3R0H1 A0A1Y1N025 A0A0N5DZH8 A0A2D1QU04 A0A2D1QU21 A0A2D1QTX3 A0A1S3MF07 A0A3B3ZGR4 H3BH40 A0A3P8ZQ78 A0A210QLV6 G3UUY2 A0A3B3ZGS3 A0A2D1QTZ2 A0A3M7S2V3 B5XAK3 B5XFI6 A0A2G8KFG3 A0A151M3K8 A0A2G8JL59 A0A3B3ZH74 V8N9T5 A0A3N0XW44 A0A3B3ZGN5
A0A212EU94 A0A194QKA5 A0A3S2NNA5 A0A194QD28 S4NYM1 A0A2H1WUT2 A0A0L7KUT3 E2AH82 A0A067QSE3 A0A087ZR76 A0A0L7QQK8 A0A1B6F003 E9IH86 A0A026WK52 A0A023F117 A0A154PHF9 A0A1B6HND8 A0A195DMZ1 A0A348G5X9 E2BFY2 A0A195BJB1 A0A158NA97 A0A0V0G611 A0A0N7Z8I4 V9IL21 A0A195FRB7 T1HUX6 A0A151X7Q4 A0A2A3E4R2 A0A0J7KMP0 D6X3Z5 A0A2K8JSP6 F4WN59 A0A224Y1P5 A0A0T6B9Q1 A0A195CBH9 A0A2J7PDN1 A0A0C9QP19 A0A1B6DJH2 A0A1W4WH03 A0A0M9ACY7 A0A1Y1LHQ0 A0A1Y1ME15 A0A146L0X7 A0A3B3R0H1 A0A1Y1N025 A0A0N5DZH8 A0A2D1QU04 A0A2D1QU21 A0A2D1QTX3 A0A1S3MF07 A0A3B3ZGR4 H3BH40 A0A3P8ZQ78 A0A210QLV6 G3UUY2 A0A3B3ZGS3 A0A2D1QTZ2 A0A3M7S2V3 B5XAK3 B5XFI6 A0A2G8KFG3 A0A151M3K8 A0A2G8JL59 A0A3B3ZH74 V8N9T5 A0A3N0XW44 A0A3B3ZGN5
PDB
1ZLG
E-value=0.00297423,
Score=89
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 28,
Likelihood: 0.996077
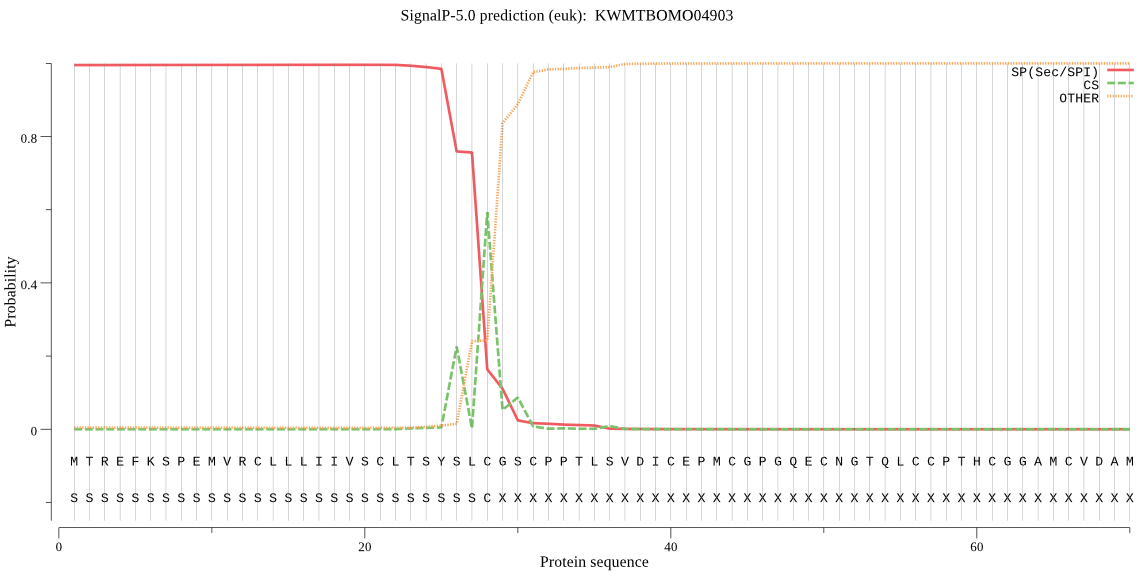
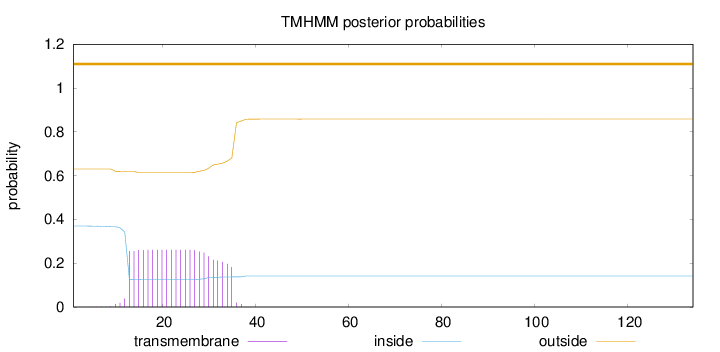
Length:
134
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.75367999999999
Exp number, first 60 AAs:
5.75305
Total prob of N-in:
0.36948
outside
1 - 134
Population Genetic Test Statistics
Pi
163.866252
Theta
130.122705
Tajima's D
0.926697
CLR
0.383805
CSRT
0.636018199090046
Interpretation
Uncertain
