Gene
KWMTBOMO04900 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008017
Annotation
aminopeptidase_N-7_[Bombyx_mori]
Full name
Aminopeptidase
+ More
Aminopeptidase N
Aminopeptidase N
Alternative Name
Apn2
Microsomal aminopeptidase
Apn1
Microsomal aminopeptidase
Apn1
Location in the cell
PlasmaMembrane Reliability : 3.796
Sequence
CDS
ATGTCAGTTTATTCATTGATCCTGCCAATACCGTCAGTTTCAACGGGAAGGAGCAGCAACGAAAACTTGTTTGCTAGTTTCACTCTAGCAACTGATGATACGCATTTTTTGAGAATCAGTACCAGAACACAGCTCCTTCCAGACCAGCCGTATATTGTGAACATTGATTATGAATCTAAATACGCGCCCAACATGTTCGGAGTATATGTATCTACATACCAACAAAATGGACGAACTGTAAATCTGGTAACTTCTCAACTTCAACCAACGTTTGCCCGCCGTGCATTCCCTTGCTACGATGAGCCGGCTATCAAAGCTATTTTCAGAACTACCATATACGCACCAGCAGCCTACACGGTTGTTAGGCACAATACTCCCGAAAGAGCTGTCCCATTGAAGGAGGACGTGGCGGGTTACGTGAAGCACGAGTTTGAAGACACCCTCGTAATGTCGACGTACCTGCTCGCCTATCTCGTGTCGAACTTCGAGCACGTTAGCAACGAGCAAAACCCCATCTACAGAGTCCCCTTCAGGGTATATTCTAGGCCAGGAACCCAGACCAACGCAGCCTTCGCAATGGACTTCGGACAAAAGAACATGGTCGCTTTGGAAGCGTACAACGAATTCCCTTATGCTTTCCCTAAATTAGATAAAGCAGCCGTTCCCGATTTTGCTGCGGGTGCCATGGAGAACTGGGGTCTTGTCATTTACAGGGAAGTGGCATTGCTGGTGACGGAAGGTGTAACGACCACAGCGACCAGGCAGAACATTGCCAGGATTATCTGCCACGAGAATGTGCACATGTGGTACGGCAATGAAGTGGGTCCGCTGTCCTGGACTTACACTTGGCTCAACGAAGGTTTCGCAACGTTCTTCGAGAGTTTCGCCACTGATTTGGTACTTCCGGAATGGCGTATGATGGAGCAGTTTGTGGTGACCATGCAGAATGTGTTCCAATCGGATGCTGTTCTCACTATCAACCCCATGACGCACGCGGTCTACACTCCTTCTCAGATCATGGGACAATTTAACGCCATTGCCTATCAAAAATCCGGCTCGGTGATCAGAATGTTGCATCATTTCCTGACACCGGAAATATTCAGACGAGGTCTCGTTATTTACATCATTAACAACTCCCGTCGTGCTGCCGGGCCGTCAGACTTATATGCTGCTCTCCAACAAGCCCTGGATGCGTCCGACCACAGCATCCCGTACTCGATCTCCAATGTGATGAACAGATGGGTCAACCAGGGAGGGTTCCCAGTGTTGAACGTGAGAAAGAGCGCCCCGAACGCTAATTCGGTCTTTATTTCTCAGGAGCGATATCTCACAGACCGCTCTCTGACGTCAACGGACCGTTGGCATGTGCCAGTCAACTGGGTGCTGTCTTCCAACATTGATTTCTCTGACACAAAACCCCAAGGTTGGATCCCACCGTCGTTCCCTGCCACCTCGATCGACATTCCTGGATTGGCAAACGCCGAATGGTTCATTTTTAACAAGCAACAGACTGGCTACTACCGCGTGAACTACGATCCGGAGAACTGGGCGGCCCTCGCAAGAGTCTTACAGACCAATCATGCCGTTATCCATCTCCTCAATCGCGCTCAGATTCTTGACGATTCCTTCAACATGGCCAGAAATGGGCGCCTGAATTACAACCTTCCCTTCGAGATTTCTCGATACCTGATCAACGAGAAAGATTACATTCCCTGGGCTGCCATTAACCCTGCCTTCAATTACTTAGATATCGTTCTAACCGGATCTTCAGTATACAATTTGTTTAGGGAGTATCTTCTGACACTGACTGCACCTCTATATGACGAAATTGGTTGGGAGGCGACTGCCAACGAGGAGCATGTTATGGCTTACCACAGAAATATCATATTGGACATCAATTGTCGTCTCGGAAACCAGAGATGCGTCACCAGAGCCCAGGAACTGTTGGAGCAGTTCCGAAATAATCCAACTCAAAGACTGAACCCGGATTTACAAAACACAGTGTACTGTTCCGGTCTACGTGGCGGTGATAGAGATAATTTCAATTTCCTTTGGGAGCAATATTTGGCTAGCTCTGATTCAAGCGAGCAAAACATCCTCCTCAATGCTCTGGGATGTACTTCGAACCCTGAACTGCGTACCTTCTACATGAACCAAGTTATCGATGCCAACTCACCAGTGAGGGAACAAGACCGGCACACAATTCTCGTCTCTGTCATCAACTCTAGTCCAGAGAACATGGACGCTGCCTTAGAATTCGTCATTGAAAACTTCCACAGGATCCAACCGAGGGTGCAAGGCCTCACCGGGACCACTAACATATTAAATGCCTTCGCAAGAAGACTGACCACAGAAACGCATGCTGAAAGGATAAACCAGCTGATCAGTCGTCACCAAGCCATCTTAACAGCTGGAGAACAAGCATCCATCTCAGCTATCAGAGAGCACATCGCAGCTTCGATAGCTTGGGGTAAAGACAATGCTGCTGTTGTAGAGGACTGGCTTGAAGACAACTACGGAGAACCCAAGCCGGAGGAACCATCCGCAGCACATTCTACGACGGCTGGTTTTATTGTTTTGCTATCTGCTTTTGTCGCTTTCTTTAACATTCATTAA
Protein
MSVYSLILPIPSVSTGRSSNENLFASFTLATDDTHFLRISTRTQLLPDQPYIVNIDYESKYAPNMFGVYVSTYQQNGRTVNLVTSQLQPTFARRAFPCYDEPAIKAIFRTTIYAPAAYTVVRHNTPERAVPLKEDVAGYVKHEFEDTLVMSTYLLAYLVSNFEHVSNEQNPIYRVPFRVYSRPGTQTNAAFAMDFGQKNMVALEAYNEFPYAFPKLDKAAVPDFAAGAMENWGLVIYREVALLVTEGVTTTATRQNIARIICHENVHMWYGNEVGPLSWTYTWLNEGFATFFESFATDLVLPEWRMMEQFVVTMQNVFQSDAVLTINPMTHAVYTPSQIMGQFNAIAYQKSGSVIRMLHHFLTPEIFRRGLVIYIINNSRRAAGPSDLYAALQQALDASDHSIPYSISNVMNRWVNQGGFPVLNVRKSAPNANSVFISQERYLTDRSLTSTDRWHVPVNWVLSSNIDFSDTKPQGWIPPSFPATSIDIPGLANAEWFIFNKQQTGYYRVNYDPENWAALARVLQTNHAVIHLLNRAQILDDSFNMARNGRLNYNLPFEISRYLINEKDYIPWAAINPAFNYLDIVLTGSSVYNLFREYLLTLTAPLYDEIGWEATANEEHVMAYHRNIILDINCRLGNQRCVTRAQELLEQFRNNPTQRLNPDLQNTVYCSGLRGGDRDNFNFLWEQYLASSDSSEQNILLNALGCTSNPELRTFYMNQVIDANSPVREQDRHTILVSVINSSPENMDAALEFVIENFHRIQPRVQGLTGTTNILNAFARRLTTETHAERINQLISRHQAILTAGEQASISAIREHIAASIAWGKDNAAVVEDWLEDNYGEPKPEEPSAAHSTTAGFIVLLSAFVAFFNIH
Summary
Catalytic Activity
Release of an N-terminal amino acid, Xaa-|-Yaa- from a peptide, amide or arylamide. Xaa is preferably Ala, but may be most amino acids including Pro (slow action). When a terminal hydrophobic residue is followed by a prolyl residue, the two may be released as an intact Xaa-Pro dipeptide.
Cofactor
Zn(2+)
Subunit
Binds to CRY1AB5.
Similarity
Belongs to the peptidase M1 family.
Keywords
Aminopeptidase
Cell membrane
Direct protein sequencing
Disulfide bond
Glycoprotein
GPI-anchor
Hydrolase
Lipoprotein
Membrane
Metal-binding
Metalloprotease
Protease
Signal
Zinc
Feature
chain Aminopeptidase
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9JES0
I3VR78
O76151
P91885
A0A286LPZ7
A0A194QLY2
+ More
Q9XYV3 B2D0I7 E8Z5D0 A3RIW4 A0A2W1BMG1 B6CMF1 Q5EFF2 B5A8Y0 A4D0J4 P91887 Q4G6A4 Q6Q2I0 D2KHN3 E0A210 A0A2H1VG23 A0A3S2LTZ5 A0A212EJU3 A0A2Z3YIN5 A0A2A4J3L5 A0A2J7RAG9 A0A2J7RAH0 A0A2J7PPF0 A0A2P8Y405 E2BB79 A0A2J7RAG5 A0A182GWW7 Q95PB9 Q8T4T6 Q16L31 U5EWR8 A0A0M9A6N6 Q95P62 A0A1S4J3H9 Q8T4T7 K7IT85 A0A0P4VSA7 A0A026W0P7 K7INE5 A0A087ZUG8 A0A3L8E4L7 A0A0M4ED13 A0A1I8P6R0 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 E2ALT1 A0A2A3EJS2 X1WUZ5 A0A1Q3FS18 A0A2S2NGP8 A0A1I8MFJ3 A0A146LYB5 A0A0V0G738 A0A0K8R857 T1P8B3 A0A232EWK0 A0A310SPN7 E0VS71 A0A2J7PPF3 A0A182GA80 A0A195F0B1 A0A1W4XT18 A0A212EJQ4 A0A2H8TGE4 B0W1Y2 A0A034V8K5 A0A2R7VTS0 A0A194QK79 A0A195BIK5 T1I7S8 A0A084WR42 A0A232FJ88 A0A0C9R2S6 B4M146 A0A1W4VQT7 A0A195DN33 B4K976 A0A1B6GWM4 A0A1W0XDI0 A0A067QSR5 E2BB86 A0A194QD38 B4NJ06 A0A195DN37 A0A182KFB7 A0A2J7RAG3 A0A0L7QWL2 B4JF64 A0A1I8P0F0 Q178P2
Q9XYV3 B2D0I7 E8Z5D0 A3RIW4 A0A2W1BMG1 B6CMF1 Q5EFF2 B5A8Y0 A4D0J4 P91887 Q4G6A4 Q6Q2I0 D2KHN3 E0A210 A0A2H1VG23 A0A3S2LTZ5 A0A212EJU3 A0A2Z3YIN5 A0A2A4J3L5 A0A2J7RAG9 A0A2J7RAH0 A0A2J7PPF0 A0A2P8Y405 E2BB79 A0A2J7RAG5 A0A182GWW7 Q95PB9 Q8T4T6 Q16L31 U5EWR8 A0A0M9A6N6 Q95P62 A0A1S4J3H9 Q8T4T7 K7IT85 A0A0P4VSA7 A0A026W0P7 K7INE5 A0A087ZUG8 A0A3L8E4L7 A0A0M4ED13 A0A1I8P6R0 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 E2ALT1 A0A2A3EJS2 X1WUZ5 A0A1Q3FS18 A0A2S2NGP8 A0A1I8MFJ3 A0A146LYB5 A0A0V0G738 A0A0K8R857 T1P8B3 A0A232EWK0 A0A310SPN7 E0VS71 A0A2J7PPF3 A0A182GA80 A0A195F0B1 A0A1W4XT18 A0A212EJQ4 A0A2H8TGE4 B0W1Y2 A0A034V8K5 A0A2R7VTS0 A0A194QK79 A0A195BIK5 T1I7S8 A0A084WR42 A0A232FJ88 A0A0C9R2S6 B4M146 A0A1W4VQT7 A0A195DN33 B4K976 A0A1B6GWM4 A0A1W0XDI0 A0A067QSR5 E2BB86 A0A194QD38 B4NJ06 A0A195DN37 A0A182KFB7 A0A2J7RAG3 A0A0L7QWL2 B4JF64 A0A1I8P0F0 Q178P2
EC Number
3.4.11.-
3.4.11.2
3.4.11.2
Pubmed
EMBL
BABH01023399
BABH01023400
BABH01023401
BABH01023402
JQ061149
AFK85023.1
+ More
AB011497 BAA32140.1 X97877 MF425654 ASU92546.1 KQ461198 KPJ05970.1 AF126443 AAD31184.1 EU564811 ACB47287.1 EU878376 ACJ64828.1 EF417486 ABN64103.1 KZ149969 PZC76118.1 EU325551 ACB54941.1 AY894814 AAW72993.1 EU826127 ACF34999.1 AY836580 AAX39864.1 X97878 AY218843 AAP44965.1 AY564237 AAS75552.1 GU213038 ADA59490.1 HM231317 ADL38969.1 ODYU01002173 SOQ39332.1 RSAL01000228 RVE43927.1 AGBW02014405 OWR41740.1 KY949254 AWT22996.1 NWSH01003448 PCG66278.1 NEVH01006570 PNF37826.1 PNF37825.1 NEVH01023278 PNF18181.1 PYGN01000952 PSN39002.1 GL446946 EFN87052.1 PNF37829.1 JXUM01094214 KQ564112 KXJ72771.1 AF378117 AAK55416.1 AY064079 AAL85580.1 KF021690 CH477924 AGT95895.1 EAT35030.1 GANO01001287 JAB58584.1 KQ435724 KOX78330.1 AF390100 AAK73351.1 AY064078 AAL85579.1 AAZX01003317 GDKW01001137 JAI55458.1 KK107589 EZA48634.1 QOIP01000001 RLU27566.1 CP012526 ALC45616.1 GBGD01000333 JAC88556.1 IACF01002668 LAB68313.1 GFTR01008371 JAW08055.1 GL440629 EFN65601.1 KZ288223 PBC31977.1 ABLF02030670 ABLF02030672 GFDL01004812 JAV30233.1 GGMR01003721 MBY16340.1 GDHC01006045 JAQ12584.1 GECL01002882 JAP03242.1 GADI01006478 JAA67330.1 KA644849 AFP59478.1 NNAY01001852 OXU22728.1 KQ760397 OAD60666.1 DS235745 EEB16227.1 PNF18179.1 JXUM01050786 JXUM01050787 JXUM01050788 JXUM01050789 KQ561662 KXJ77885.1 KQ981905 KYN33597.1 OWR41732.1 GFXV01001305 MBW13110.1 DS231824 EDS27418.1 GAKP01020867 JAC38085.1 KK854013 PTY09335.1 KPJ05978.1 KQ976467 KYM84197.1 ACPB03005577 ATLV01025918 KE525402 KFB52686.1 NNAY01000153 OXU30528.1 GBYB01001141 JAG70908.1 CH940650 EDW67457.1 KQ980713 KYN14242.1 CH933806 EDW14489.1 GECZ01002996 JAS66773.1 MTYJ01000002 OQV25529.1 KK853596 KDR06435.1 EFN87059.1 KQ459185 KPJ03377.1 CH964272 EDW84908.1 KYN14241.1 PNF37828.1 KQ414710 KOC62990.1 CH916369 EDV93345.1 CH477360 EAT42699.1
AB011497 BAA32140.1 X97877 MF425654 ASU92546.1 KQ461198 KPJ05970.1 AF126443 AAD31184.1 EU564811 ACB47287.1 EU878376 ACJ64828.1 EF417486 ABN64103.1 KZ149969 PZC76118.1 EU325551 ACB54941.1 AY894814 AAW72993.1 EU826127 ACF34999.1 AY836580 AAX39864.1 X97878 AY218843 AAP44965.1 AY564237 AAS75552.1 GU213038 ADA59490.1 HM231317 ADL38969.1 ODYU01002173 SOQ39332.1 RSAL01000228 RVE43927.1 AGBW02014405 OWR41740.1 KY949254 AWT22996.1 NWSH01003448 PCG66278.1 NEVH01006570 PNF37826.1 PNF37825.1 NEVH01023278 PNF18181.1 PYGN01000952 PSN39002.1 GL446946 EFN87052.1 PNF37829.1 JXUM01094214 KQ564112 KXJ72771.1 AF378117 AAK55416.1 AY064079 AAL85580.1 KF021690 CH477924 AGT95895.1 EAT35030.1 GANO01001287 JAB58584.1 KQ435724 KOX78330.1 AF390100 AAK73351.1 AY064078 AAL85579.1 AAZX01003317 GDKW01001137 JAI55458.1 KK107589 EZA48634.1 QOIP01000001 RLU27566.1 CP012526 ALC45616.1 GBGD01000333 JAC88556.1 IACF01002668 LAB68313.1 GFTR01008371 JAW08055.1 GL440629 EFN65601.1 KZ288223 PBC31977.1 ABLF02030670 ABLF02030672 GFDL01004812 JAV30233.1 GGMR01003721 MBY16340.1 GDHC01006045 JAQ12584.1 GECL01002882 JAP03242.1 GADI01006478 JAA67330.1 KA644849 AFP59478.1 NNAY01001852 OXU22728.1 KQ760397 OAD60666.1 DS235745 EEB16227.1 PNF18179.1 JXUM01050786 JXUM01050787 JXUM01050788 JXUM01050789 KQ561662 KXJ77885.1 KQ981905 KYN33597.1 OWR41732.1 GFXV01001305 MBW13110.1 DS231824 EDS27418.1 GAKP01020867 JAC38085.1 KK854013 PTY09335.1 KPJ05978.1 KQ976467 KYM84197.1 ACPB03005577 ATLV01025918 KE525402 KFB52686.1 NNAY01000153 OXU30528.1 GBYB01001141 JAG70908.1 CH940650 EDW67457.1 KQ980713 KYN14242.1 CH933806 EDW14489.1 GECZ01002996 JAS66773.1 MTYJ01000002 OQV25529.1 KK853596 KDR06435.1 EFN87059.1 KQ459185 KPJ03377.1 CH964272 EDW84908.1 KYN14241.1 PNF37828.1 KQ414710 KOC62990.1 CH916369 EDV93345.1 CH477360 EAT42699.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000007151
UP000218220
UP000235965
+ More
UP000245037 UP000008237 UP000069940 UP000249989 UP000008820 UP000053105 UP000002358 UP000053097 UP000005203 UP000279307 UP000092553 UP000095300 UP000000311 UP000242457 UP000007819 UP000095301 UP000215335 UP000009046 UP000078541 UP000192223 UP000002320 UP000078540 UP000015103 UP000030765 UP000008792 UP000192221 UP000078492 UP000009192 UP000027135 UP000053268 UP000007798 UP000075881 UP000053825 UP000001070
UP000245037 UP000008237 UP000069940 UP000249989 UP000008820 UP000053105 UP000002358 UP000053097 UP000005203 UP000279307 UP000092553 UP000095300 UP000000311 UP000242457 UP000007819 UP000095301 UP000215335 UP000009046 UP000078541 UP000192223 UP000002320 UP000078540 UP000015103 UP000030765 UP000008792 UP000192221 UP000078492 UP000009192 UP000027135 UP000053268 UP000007798 UP000075881 UP000053825 UP000001070
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JES0
I3VR78
O76151
P91885
A0A286LPZ7
A0A194QLY2
+ More
Q9XYV3 B2D0I7 E8Z5D0 A3RIW4 A0A2W1BMG1 B6CMF1 Q5EFF2 B5A8Y0 A4D0J4 P91887 Q4G6A4 Q6Q2I0 D2KHN3 E0A210 A0A2H1VG23 A0A3S2LTZ5 A0A212EJU3 A0A2Z3YIN5 A0A2A4J3L5 A0A2J7RAG9 A0A2J7RAH0 A0A2J7PPF0 A0A2P8Y405 E2BB79 A0A2J7RAG5 A0A182GWW7 Q95PB9 Q8T4T6 Q16L31 U5EWR8 A0A0M9A6N6 Q95P62 A0A1S4J3H9 Q8T4T7 K7IT85 A0A0P4VSA7 A0A026W0P7 K7INE5 A0A087ZUG8 A0A3L8E4L7 A0A0M4ED13 A0A1I8P6R0 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 E2ALT1 A0A2A3EJS2 X1WUZ5 A0A1Q3FS18 A0A2S2NGP8 A0A1I8MFJ3 A0A146LYB5 A0A0V0G738 A0A0K8R857 T1P8B3 A0A232EWK0 A0A310SPN7 E0VS71 A0A2J7PPF3 A0A182GA80 A0A195F0B1 A0A1W4XT18 A0A212EJQ4 A0A2H8TGE4 B0W1Y2 A0A034V8K5 A0A2R7VTS0 A0A194QK79 A0A195BIK5 T1I7S8 A0A084WR42 A0A232FJ88 A0A0C9R2S6 B4M146 A0A1W4VQT7 A0A195DN33 B4K976 A0A1B6GWM4 A0A1W0XDI0 A0A067QSR5 E2BB86 A0A194QD38 B4NJ06 A0A195DN37 A0A182KFB7 A0A2J7RAG3 A0A0L7QWL2 B4JF64 A0A1I8P0F0 Q178P2
Q9XYV3 B2D0I7 E8Z5D0 A3RIW4 A0A2W1BMG1 B6CMF1 Q5EFF2 B5A8Y0 A4D0J4 P91887 Q4G6A4 Q6Q2I0 D2KHN3 E0A210 A0A2H1VG23 A0A3S2LTZ5 A0A212EJU3 A0A2Z3YIN5 A0A2A4J3L5 A0A2J7RAG9 A0A2J7RAH0 A0A2J7PPF0 A0A2P8Y405 E2BB79 A0A2J7RAG5 A0A182GWW7 Q95PB9 Q8T4T6 Q16L31 U5EWR8 A0A0M9A6N6 Q95P62 A0A1S4J3H9 Q8T4T7 K7IT85 A0A0P4VSA7 A0A026W0P7 K7INE5 A0A087ZUG8 A0A3L8E4L7 A0A0M4ED13 A0A1I8P6R0 A0A069DXK6 A0A2P2I2Q6 A0A224X6F7 E2ALT1 A0A2A3EJS2 X1WUZ5 A0A1Q3FS18 A0A2S2NGP8 A0A1I8MFJ3 A0A146LYB5 A0A0V0G738 A0A0K8R857 T1P8B3 A0A232EWK0 A0A310SPN7 E0VS71 A0A2J7PPF3 A0A182GA80 A0A195F0B1 A0A1W4XT18 A0A212EJQ4 A0A2H8TGE4 B0W1Y2 A0A034V8K5 A0A2R7VTS0 A0A194QK79 A0A195BIK5 T1I7S8 A0A084WR42 A0A232FJ88 A0A0C9R2S6 B4M146 A0A1W4VQT7 A0A195DN33 B4K976 A0A1B6GWM4 A0A1W0XDI0 A0A067QSR5 E2BB86 A0A194QD38 B4NJ06 A0A195DN37 A0A182KFB7 A0A2J7RAG3 A0A0L7QWL2 B4JF64 A0A1I8P0F0 Q178P2
PDB
4WZ9
E-value=1.62901e-122,
Score=1128
Ontologies
PATHWAY
GO
PANTHER
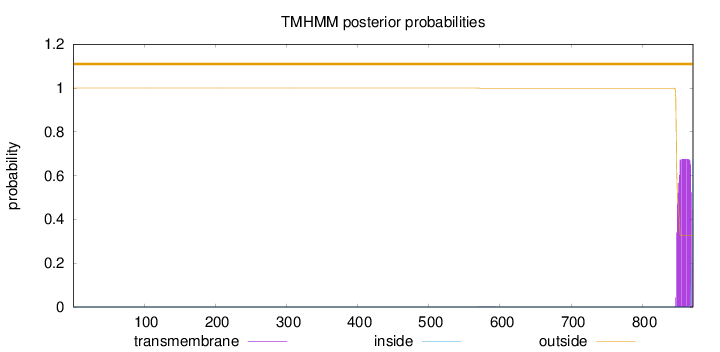
Topology
Subcellular location
Cell membrane
Length:
871
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.29196
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.00007
outside
1 - 871
Population Genetic Test Statistics
Pi
241.413237
Theta
169.520407
Tajima's D
1.532915
CLR
0.329373
CSRT
0.7961601919904
Interpretation
Uncertain
